Introduction
Kefir is a fermented milk beverage that is known to have originated in the Caucasus (Liu and Lin, 2000; Fontán et al., 2002). The word “kefir” is derived from the Turkish word keyif, which means pleasure/pleasurable. The drink is also produced in countries such as Argentina, Taiwan, Portugal, France and Turkey under different names such as "kephir", "kiaphur", "kefer", "kefyr", "knapon", "kepi" and "kippi" (Leite et al., 2015). Kefir is produced by fermenting milk with kefir grain, which has a complex microflora and can be re-used in the fermentation of the next batch of kefir (Farnworth, 2006). Kefir grain contains lactic acid bacteria (lactobacilli, lactococci, streptococci, leuconostoc), yeast and acetic acid bacteria (Marshall and Cole, 1985; Liu and Lin, 2000; Powell et al., 2007; Pogačić et al., 2013). The microorganisms most commonly isolated from kefir grain and beverages include Lactobacillus kefiranofaciens Lb. buchneri, Lb. helveticus, Lb. kefiri, Lb. acidophilus, Lb. casei, and Lactococcus lactis (Kesmen and Kaçmaz, 2011; Nalbantoğlu et al., 2014; Dertli and Çon, 2017). Koumiss (airag, chige, arrag or chigo in Mongolian) is a fermented alcoholic milk beverage that is produced from mare's milk and is consumed in Central Asia especially in Mongolia (Danova et al., 2005; Akuzawa et al., 2011). Microorganisms involved in koumiss fermentation often include lactobacilli such as, Lb. salivarius, Lb. buchneri, Lb. plantarum (Danova et al., 2005), Lb. delbrueckii subsp. bulgaricus (Akuzawa et al., 2011), Lb. helveticus, Lb. fermentum (Wang et al., 2008), L. casei (Ya et al., 2008), Lb. kefiranofaciens (Watanabe et al., 2008). In Turkey koumiss production and consumption is limited, so there is limited research on koumiss microflora. Until recent years cultural and nucleic acid-based methods have been used in the detection of bacterial microflora of kefir and koumiss, whereas metagenomic analysis have been frequently preferred in last years (Gao et al., 2013; Nalbantoğlu et al., 2014; Dertli and Çon, 2017; Yao et al., 2017). Recently, the importance of the metagenomics approaches in the food analysis is increasing (Walsh et al., 2017). The microbial species found in foods or that contaminate them have been identified using cultural methods, and these techniques have proven their effectiveness in shaping the rules and regulations related to food safety. However, these cultural approaches have the disadvantage of detecting only the culturable microorganisms within the real microbial population (Giraffa and Neviani, 2011). The development of new generation sequencing technologies and their application in the food microbiology area has shown that microbial diversity in these products is in fact greater than initially assumed. While the use of new generation sequencing techniques is relatively new in food microbiology, their popularity is gradually increasing, and their use has started to attract the attention of not only researchers, but also many food producers (Kergourlay et al., 2015). Probiotics were defined by the World Health Organization as living microorganisms that have positive effects on the consumer health when they are consumed sufficiently (FAO/WHO, 2001). To provide health benefits, probiotics must overcome physical and chemical barriers such as acid and bile in the gastrointestinal tract (Bao et al., 2010; Kos et al., 2011). To survive and carry out their physiological activities during their passage through the gastrointestinal tract, probiotic microorganisms must be resistant to the acidic environment of the stomach and the bile salts of the intestines. For this reason, acid and bile salt resistance is considered to be the main criteria in the identification of probiotic strains.
The objective of this study was to determine bacterial diversity of kefir and koumiss beverages consumed in Turkey and comparing the probiotic properties (acid and bile salt resistance) of lactic acid bacteria isolated from these beverages with Lb. casei used in yakult production.
Materials and methods
Sample collection
In this study we used one koumiss and 5 different commercially available kefir beverages with different production dates and one homemade kefir used as samples. Homemade kefir was produced by grains obtaining from a family, producing kefir for their own consumption. 20 g of grain inoculated into 500 mL UHT milk (3 % fat), after which the mixture was incubated at room temperature (about 25 ºC) for 12 hours for activation of grains. At the end of the incubation period, the grains were filtered and inoculated again into 500 mL of UHT milk, and allowed to incubate once again. The obtained kefir beverage was used as a sample.
For cultural isolation, 1 mL sample was diluted in 10 mL ¼ strength Ringer (Merck, Germany) solution and spread on to MRS (de Man, Rogosa and Sharpe) (Merck, Germany) and M17 (Merck, Germany) agar media. MRS agar plates were incubated at 37 ºC for 72 hours in anaerobic jars with Anaerocult C (Merck,Germany), and M17 agar plates were incubated at 37 ºC for 48-72 hours under aerobic conditions. The milky white colonies that grew on the petri dishes were counted and then transferred into tubes for DNA isolation.
DNA isolation
DNA was isolated from all of the colonies that grew on the MRS and M17 agar, and directly from the food samples. DNA isolation was performed by a protocol designed for total DNA from gram-positive bacteria using a commercially available genomic DNA purification kit (DNeasy PowerFood Microbial Kit, Qiagen). DNA concentrations were determined using a nanodrop spectrophotometer (Titertek Berthold, Germany). Following this, electrophoresis was run, using 1 % agarose gel in an electrophoresis tank containing a 1X Tris Acetic EDTA (TAE) buffer solution. The obtained electrophoresis (Thermo Scientific OWL EasyCast B2, USA) results were confirmed through visualization with a UV Transilluminator (DNR Bio-Imaging Systems MiniBIS Pro, Israel).
Metagenome analysis
DNA isolation was made in triplicate for each sample to get more diversity. The DNA that was isolated from the colonies grown on the MRS and M17 agar and directly from the beverages were pooled in a way that there were three tubes of each sample (each three tubes containing DNA from the MRS agar, M17 agar, and directly from beverages, respectively). Samples were sequenced on Illumina MiSeq platform in BMLabosis (Ankara, Turkey). The V1-V2 hypervariable region of the 16S rRNA gene was amplified using specific primers. The forward primer was AGAGTTTGATCMTGGCTCAG and the reverse primer was CYIACTGCTGCCTCCCGTAG. For indexing, a PCR reaction was done with an initial denaturation of 3 min at 95 ºC; 8 cycles of 30 sec at 95 ºC, 30 sec at 55 ºC and 30 sec at 72 ºC; and a post elongation of 5 min at 72 ºC. This PCR reaction was performed using the Nextera XT index kit (FC-131-1001/FC-131-1002) for Illumina sequencing protocols.
After the sequencing, QIIME 2 software was used for the determination of taxonomic species (Bolyen et al., 2018). Alpha diversity analysis was performed to determine the species within a sample, and beta diversity analysis was performed to detect the diversity or similarity between samples.
Investigation of probiotic properties
Among the colonies grown on the MRS and M17 agar, 23 colonies were selected according to the colony morphology and re-inoculated in MRS and M17 agar. The growing colonies were stained according to Gram and analyzed for catalase activity.
Tests were carried out on the Gram-positive and catalase negative colonies to identify their probiotic properties.
Measurement of acid tolerance
The acid tolerance test was carried out combining the methods of Charteris et al. (1998) and Klingber et al. (2005). The cultures inoculated in the MRS and M17 broths were incubated until their optical density reached 0.6 at a 600 nm on a spectrophotometer (Eppendorf BioPhotometer, Germany), (corresponding to approximately 10 9 cells/mL). Following this, an inoculation was performed (1 % v/v) into MRS and M17 broths with pH values brought to 2.5 using HCl (Merck, Germany), and viability at the start after and after 2 hours was determined through spreading on the MRS and M17 agar.
Measurement of bile salt tolerance
A bile salt tolerance test was carried out, using a method recommended by Fernandez et al. (2003) with slight modification. The cultures that had been grown in the MRS and M17 broths until their OD values reached 0.6 at 600 nm (corresponding to approximately 10 9 cells/mL) were inoculated (1% v/v) into 10 mL MRS and M17 broths containing 0.3 % Oxgall (Bovine Bile, Sigma-Aldrich, Germany), and their viability at 0 th, 90 th and 180 th minutes were inspected through spreading on the MRS and M17 agar.
Identification of the selected autochthonous lactic acid bacteria
Colonies showed resistance to acids and bile salt. DNA was isolated from the colonies grown on the MRS and M17 agar using a DNeasy PowerFood Microbial Kit (DNeasy PowerFood Microbial Kit, Qiagen). The A 260/A 280 and A 260/A 230 ratios of the isolated DNAs were determined using a nanodrop spectrophotometer (Titertek Berthold, Germany) and the DNA samples stained with 2 µL of 6X loading dye (Fermentas, USA) were run in 1 % agarose gel for 60 minutes at 120 V and 100 A inside a gel tank (Thermo Scientific OWL EasyCast B2, USA) connected to a power unit (Thermo Scientific EC 300 XL, USA). The quality of the obtained DNA bands was evaluated visually on a UV transluminator (DNR Bio-Imaging Systems MiniBIS Pro, Israel). Lyophilized 27F and 1492R primers were first dissolved in ddH2O at a concentration of 100 pmol/µL. Using 1 µL of MyTaq HS DNA Polymerase (Bioline, UK) enzyme and 10 µL of a MyTaq reaction buffer (5X buffer; 5 mM dNTP, 15 mM MgCl2, stabilizers and enhancers), these 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5′-TACGGYTACCTTGTTACGACTT-3') primers (Abushelaibi et al., 2017) were mixed such that there would be 2 µL DNA, 0.1 µL 25 pmol forward primer and 0.1 µL 25 pmol reverse primer in each tube, after which they were amplified. The thermal cycler conditions of PCR reaction was consisted of an initial denaturation of 2 min at 94 ºC; 35 cycles of 2 min at 94 ºC, 20 sec at 53 ºC and 1,5 min at 70 ºC; and a post elongation of 5 min at 70 ºC in a thermal cycler (Bioneer MyGenie 96 Thermal Block-Cycler, Korea) (Ayyash et al., 2018).
The resulting PCR products were transported to a laboratory (BMLabosis, Ankara, Turkey) for duplex sequencing, under cold chain, and the sequencing results obtained from the isolates were compared with the 16S rRNA sequences found on the NCBI database (NCBI BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the closest species was identified. The interspecies phylogenetic proximity was determined using the MEGA X program.
Statistical analysis
The statistical analysis was performed to determine the differences in probiotic properties of isolates. One-way analysis of variance (ANOVA) was performed with Duncan's multiple range test post-hoc comparison. p<0.05 was considered statistically significant. Statistical analysis was performed by a statistics software package program (SPSS 20.0 IBM Corp. Released 2011. IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp.).
Results and discussion
The present study intended to determine microbiota of commercially available kefir and koumiss and homemade kefir using metagenomic analysis. In addition, it was aimed the detection of acid and bile tolerances of lactic acid bacteria isolated from these beverages.
Diversity analysis
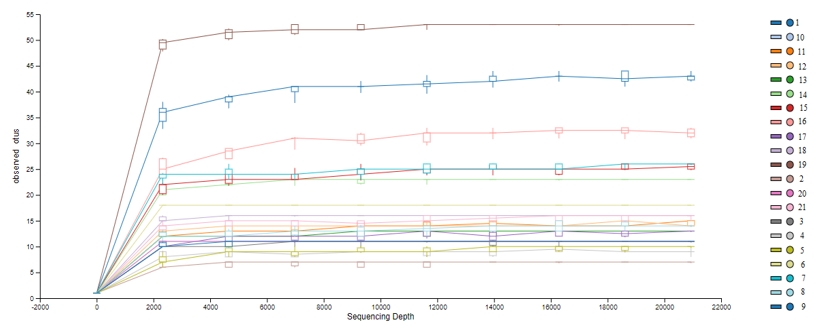
The operational taxonomic units (OTUs) obtained from DNAs isolated from MRS and M17 agars and directly from beverages were shown in Figure 1. According to OTU amounts Shannon index values (Figure 2) was calculated for determine the alpha diversity of samples.
Koumiss (1: DNA from direct beverage, 8: DNA from MRS agar, 15: DNA from M17 agar)
Kefir 1 (2: DNA from direct beverage, 9: DNA from MRS agar, 16: DNA from M17 agar)
Kefir 2 (3: DNA from direct beverage, 10: DNA from MRS agar, 17: DNA from M17 agar)
Kefir 3 (4: DNA from direct beverage, 11: DNA from MRS agar, 18: DNA from M17 agar)
Kefir 4 (5: DNA from direct beverage, 12: DNA from MRS agar, 19: DNA from M17 agar)
Kefir 5 (6: DNA from direct beverage, 13: DNA from MRS agar, 20: DNA from M17 agar)
Homemade kefir (7: DNA from direct beverage, 14: DNA from MRS agar, 21: DNA from M17 agar)
Figure 1. OTU graph. The lines parallel to the right in the α diversity graph indicate that the number of readings is sufficient for analysis
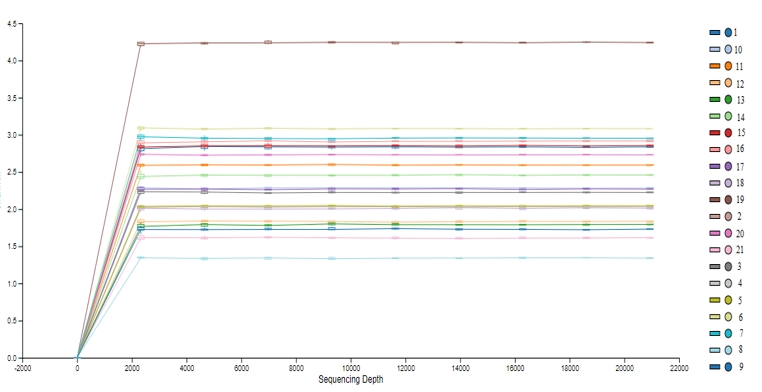
Figure 2. Shannon index values. The high Shannon index value indicates the abundance of diversity within the sample
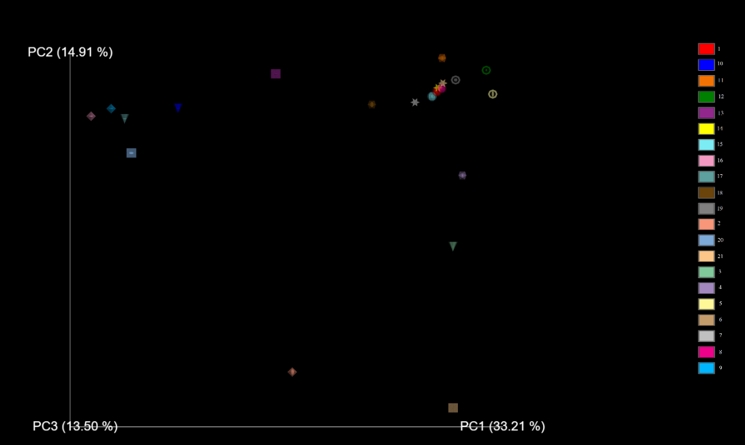
Figure 3. PCoA (principle coordinate analysis) to show the β- diversity of samples. Same shapes (sphere, diamond, cone, icosahedron, ring, square, star) indicates the same samples’ DNA extractions from direct beverages, MRS agar and M17 agar. (Sphere: Koumiss, Diamond: Kefir 1, Cone: Kefir 2, Icosahedron: 3 Ring: Kefir 4, Square: Kefir 5, Star: Homemade kefir)
In the beta diversity analysis (Figure 3) homemade kefir samples and koumiss have shown high similarity. Lb. kefiranofaciens was found to be dominant bacteria in these two samples. Number 4 and 5 kefir beverages were very close to homemade kefir and koumiss beverages. MRS and M17 agar media variety of kefir beverage number 3 was found to be far from the variety obtained directly from beverages. The agar medium diversity of kefir beverages 1,2 and 5 was observed to be close to each other. However, the diversity obtained as a result of DNA extraction directly from these beverages was determined different on the PCoA graph.
Microbial biodiversity of beverages
The dominant flora of the commercially available kefir beverages had Lc. lactis and Str. thermophilus; and the dominant species grown in MRS and M17 agar had Lc. lactis, although Leu. mesenteroides was dominant in sample three and five. Lb. kefiranofaciens was dominant in direct extraction from kefir produced using kefir grains, while Lb. kefir, Leu. mesenteroides, E. faecium and E. durans were the dominant species in MRS and M17 agars. Leu. mesenteroides was identified at low levels in the samples directly from kefir beverages, but the MRS agar showed high levels of Leu. mesenteroides. Enterococcus species could not be detected in the samples directly from the kefir beverages produced from kefir grain, although they were detected in the samples from the M17 agar. The dominant flora in the koumiss samples was Lb. kefiranofaciens, although Lb. delbrueckii was also detected. On the other hand, the dominant species that grew in the agar were Lb. kefiri, E. durans, Str. thermophilus and Massilia alkalitolerans. Lactobacillus kefiri, E. durans or Str. thermophilus could not be detected in the samples directly from koumiss beverages, but were identified in the cultural isolations. The bacterial microbiota of kefir and koumiss beverages is shown in Figure 4.
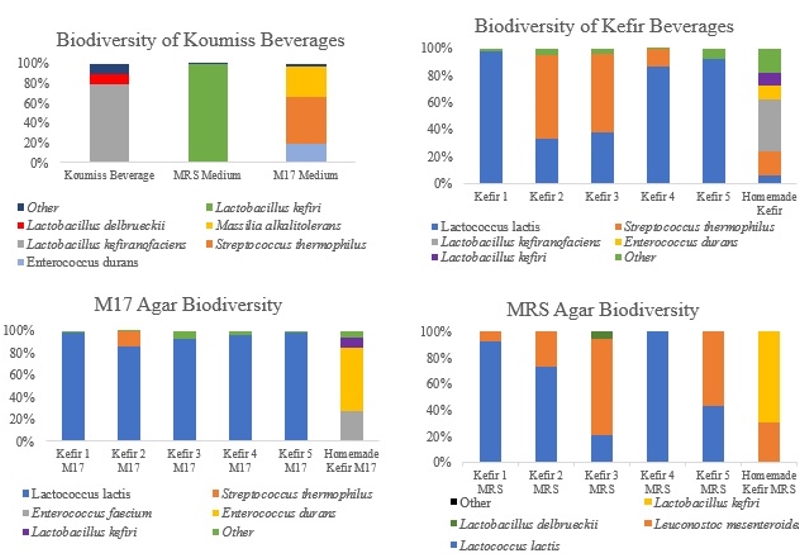
Figure 4. Microbial biodiversity of lactic acid bacteria population of kefir and koumiss beverages
The diversity between the samples from direct beverages and colonies grew on agars were different. This is thought to be due to extraction difficulties arising from the inhibitory compounds of food matrix (lipids, proteins, polysaccharides etc.) and promoting growth effect of selective mediums on injured bacteria.
Denaturing Gradient Gel Electrophoresis (PCR-DGGE) was the most frequently used culture independent method for analysis of kefir grains microbial population (Pogačić et al., 2013). A study by Kesmen and Kaçmaz (2011) clarified the bacterial population of three kefir grains and four kefir beverages collected from various regions of Turkey through the use of both culture-independent and cultural methods. The V3 region of the 16S rRNA gene was analysed using a PCR-DGGE approach, and a total of 10 different bacterial species were reported. The researchers found that the dominant flora in the kefir grain was Lb. kefiranofaciens, while the dominant flora in the kefir beverages was Lc. lactis, which concurs with the findings of the present study. The most common species were isolated using the culture-dependent technique due to the decrease on the prevalence of Lc. lactis, Leu. mesenteroides and Lb. kefiri. A review of previous studies of kefir reveals that most studies were focused on investigating the microbial diversity of kefir grain, while only few publications reported studies on kefir beverages. The findings of this study performed on kefir beverages sold in Turkey with the data in the present study. In a similar study conducted by Nalbantoğlu et al. (2014) detailed the microflora of two Turkish kefir grain samples through the use of the whole genome shotgun pyrosequencing method, and found Lactobacillus to be the most common genus, with Lb. kefiranofaciens, Lb. buchneri and Lb. helveticus species being reported as the dominant flora. Aside from these, species such as Lb. acidophilus, Lb. sunkii, Lb. johnsonii, Lb. crispatus, Lb. kefiri, Lb. casei, Lb. plantarum and Lb. delbrueckii have been identified in lower quantities. The microbial population of kefir includes a wide variety of bacterial groups, and these microorganisms are held together by a water-insoluble polysaccharide material. It has been observed that while the microbial diversity of the grain collected from different countries and regions around the world tends to be different, generally, the dominant bacteria in the media are those of the Lactobacillus genus. The species of this genus that have been most frequently identified in studies are Lb. kefiranofaciens and Lb. kefiri, while in the present study the microflora of the kefir beverages produced using kefir grain also contained Lb. kefiranofaciens. In a study conducted by Dertli and Çon (2017) using a pyrosequencing method, the microbial flora of four different kefir grains obtained from different regions was identified, and demonstrated the effects of these microorganisms on the kefir aroma. The study identified Lb. kefiranofaciens as having the largest population, while other bacteria such as Enterobacter amnigenus, Enterobacter hormaechei, Lb. kefiri, Lb. apis and Lb. ultunensis were also identified in the flora. The present study also revealed that the microbiota of the kefir samples produced from kefir grain was dominated by Lb. kefiranofaciens and Lb. kefiri. Dobson et al. (2011) determined the microbial diversity of kefir grain obtained from Ireland and of the kefir beverages produced from this grain. According to their results, the inner and outer parts of the kefir grain were formed by the Lactobacillaceae family bacteria at ratios of 88 % and 96 % respectively, while the identified species were reported as Lb. kefiranofaciens, Lb. kefiri, Lb. parabuchneri, Lb. kefiranofaciens ssp. kefirgranum, Lb. helveticus, Lb. acidophilus and Lb. parakefiri. Interestingly, the same study described that while Streptococcaceae was detected at very low levels in kefir grain, it was the dominant family in kefir beverages. Researchers have described that while Lc. lactis shows low adherence to kefir grain, it is the dominant flora in milk owing to its high competitiveness. A study conducted by Hao et al. (2010) determined the microbial populations of 10 koumiss samples collected from different regions of China using the PCR-DGGE technique. Based on the results of their study, Lb. acidophilus, Lb. helveticus, Lb. fermentum and Lb. kefiranofaciens were reported as the most dominant species, while E. faecalis, Lc. lactis, Lb. paracasei, Lb. kitasatonis and Lb. kefiri were reported to be detected to a lesser extent, along with Leu. mesenteroides, Str. thermophilus, Lb. buchneri and Lb. jensenii, albeit at much lower levels. Guo et al. (2019) investigated the microbial population of 11 different koumiss samples obtained from three different regions of Mongolia. Following the amplification of the 16S rRNA region, Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, Deinococcus-Thermus and Cyanobacteria phyla were detected, with Firmicutes being the most common among them. Lc. lactis, Lb. buchneri, Enterococcus italicus, Lb. homohiochii, Lb. hilgardii, Lb. helveticus, Leu. mesenteroides and Str. parauberis have been reported as the most commonly identified species.
Specific functional properties of selected lactic acid bacteria
Tests were performed on lactic acid bacteria isolated from beverages in order to determine the acid and bile salt resistance profiles, which are the main probiotic properties. The resistance profiles of lactic acid bacteria isolated from MRS and M17 agars are shown in Tables 1-4. Only isolate 6 has shown an increase (0,23 log 10/mL) during 2 hours of incubation at pH 2,5. Other isolates have shown a decrease ranged from 0,03-2,49 log 10/mL. During the first 90 minutes of incubation in %0,3 ox gall, concentration isolate 14 has shown the highest increase (0,71 log 10/mL). Between the 90 th and 180 th minutes, isolate 4 had the highest increase (0,73 log 10/mL).
Table 1. Acid resistance profile of M17 agar colonies (log 10/mL)
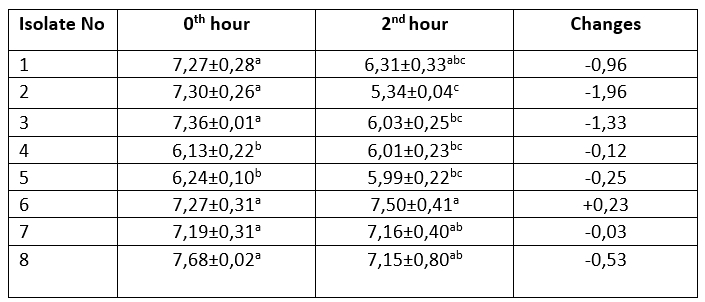
The mean ± SE values of bacterial counts (log 10 CFU/mL) on sampling hours. Refers to significant differences among isolates (Duncan's Multiple Range Test, p<0.05, after adjustment for multiple comparisons). Different letters in the same column are statistically significant (p<0.05)
Table 2. Acid resistance profile of MRS agar colonies (log 10/mL)
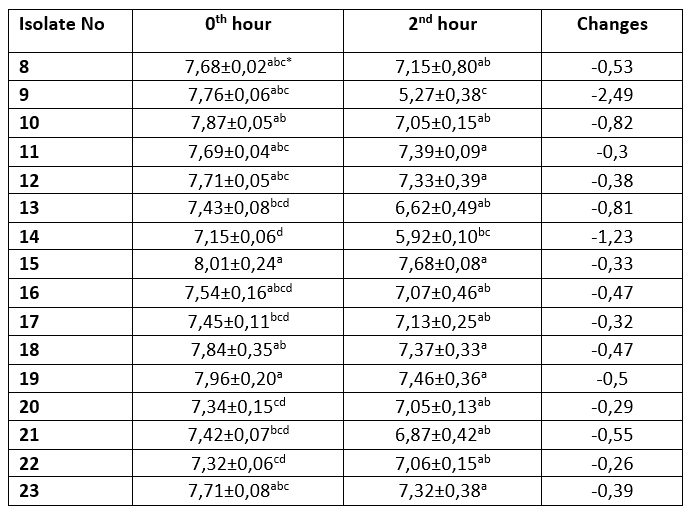
The mean ± SE values of bacterial counts (log10 CFU/mL) on sampling hours. Refers to significant differences among isolates (Duncan's Multiple Range Test, p<0.05, after adjustment for multiple comparisons). Different letters in the same column are statistically significant (p < 0.05)
Table 3. Bile salt resistance profile of M17 agar colonies (log 10/mL)
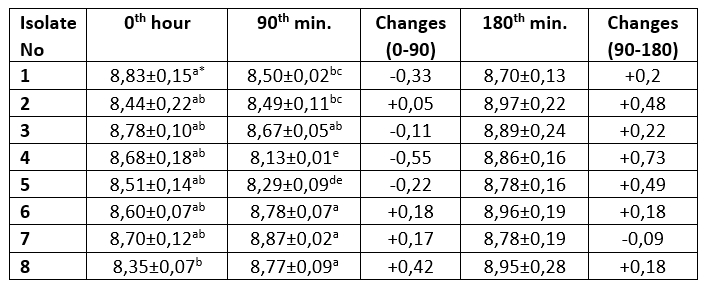
The mean ± SE values of bacterial counts (log 10 CFU/mL) on sampling hours. Refers to significant differences among isolates (Duncan's Multiple Range Test, p<0.05, after adjustment for multiple comparisons). Different letters in the same column are statistically significant (p<0.05)
Table 4. Bile salt resistance profile of MRS agar colonies (log 10/mL)
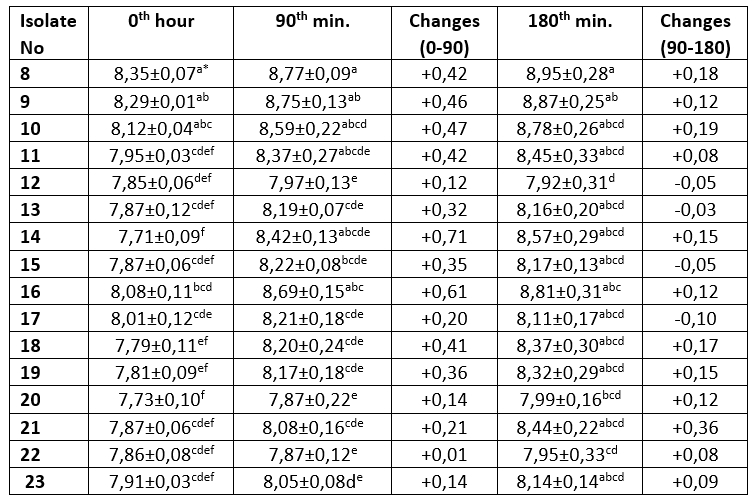
The mean ± SE values of bacterial counts (log10 CFU/mL) on sampling hours. Refers to significant differences among isolates (Duncan's Multiple Range Test, p<0.05, after adjustment for multiple comparisons). Different letters in the same column are statistically significant (p<0.05)
It is one of the main desirable characteristics of lactic acid bacteria, especially used as starter cultures, to produce lactic acid and maintain their viability in acidic environment (Leboš Pavunc et al., 2013; Hazır Dalca et al., 2018). Lactic acid bacteria are able to survive in high-acid foods without significant losses in the number of viable microorganisms (Liu et al., 2011). Although there were statistically significant differences compared to Lb. casei from yakult, all tested isolates from kefir and koumiss showed high resistance to acid and bile salts in the current study. The survival rates of Lactobacillus species at low pH levels have been found to differ considerably in different studies. In the study by Charteris et al. (1998), for example, Lb. delbrueckii subsp. bulgaricus was reported to have a low survival rate in acidic environments. In contrast to that, Vinderola and Reinheimer (2003) reported that Lb. delbrueckii remained highly viable at a pH level of 2. These results suggest that acid resistance may vary even among the same species of lactic acid bacteria. Ren et al. (2014) reported that all of the tested lactic acid bacterial strains remained highly viable at a pH of 2, and that there was no significant decrease in the number of viable bacteria at a pH of 3. In present study, in an evaluation of the resistance to bile salt of colonies grown on M17 agar, isolates number 1, 3, 4 and 5 showed a decrease in count ranging from 0.11 to 0.55 log 10/mL, while the Lb. casei isolate, number 8, isolated from the yakult showed the highest increase in count by 0.42 log 10/mL. The bacteria isolated from MRS agar showed that all isolates exhibited an increase in the first 90 minutes. The greatest increase (0.71 log 10/mL) was observed in Lb. plantarum isolate number 14, while the lowest increase was identified (0.01 log 10/mL) in Lb. kefiri sample number 22. Between 90th and 180 th minutes, isolate numbers 12, 13, 15 and 17 recorded a decrease of between 0.03-0.1 log 10/mL, while all other isolates were found to increase (0.08-0,36 log 10/mL). The Lb. casei isolate obtained from the yakult showed an increase of 0.42 log 10/mL in 90 minutes, and an increase of 0.18 log 10/mL between the 90 th and 180 th minutes.
The concentration of bile in humans is known to be physiologically between 0.3 and 0.5 %. The bile tolerance of lactic acid bacteria strains is highly important in the selection of probiotic strains, as the more resistant a strain is to bile salts, the more effective it will be in alleviating the symptoms of lactose intolerance (Vinderola and Reinheimer, 2003; Ren et al., 2014). In a similar study conducted by Ren et al. (2014) it was reported that all of the strains survived in media containing bile concentrations approximately three times higher (1 %) than the concentration observed in humans. The researchers described that they identified Lb. casei subsp. casei, Lb. delbrueckii subsp. bulgaricus, Lb. salivarius subsp. salicinius, Lb. fermentum, Lb. acidophilus, Lb. plantarum and Lb. rhamnosus bacteria from yogurt, cheese, milk, vegetables and the intestines. Sabir et al. (2010) conducted a study in which they investigated the viability of different pH and bile salt concentrations of Lb. casei subsp. casei, Lb. delbrueckii subsp. bulgaricus, Lb. salivarius subsp. salicinius, Lb. fermentum, Lb. acidophilus, Lb. plantarum and Lb. rhamnosus isolated from homemade kefir samples. In the present study, we determined that the bacteria tested at the pH and bile salt levels used in this study (2.5 and 0.3 %, respectively) maintained their viability at different levels. Similar to the results of the aforementioned studies, the present study demonstrated that Lc. lactis subsp. lactis and Lc. lactis subsp. cremoris maintained their viabilities. A study conducted by Doğan and Ozpinar (2017) investigated the probiotic properties of 130 isolates obtained from boza, cheese, kefir and raw milk samples. Based on the obtained results, it could be determined that the three kefir isolates exhibited probiotic properties in terms of hydrophobicity and resistance to acid and bile salts. These three isolates were identified using MALDI-ToF MS and were reported to be Lb. plantarum. In the present study, the sequencing of the 16S rRNA region of the colonies isolated from the MRS agar identified Lb. plantarum (10 isolates), Lb. rhamnosus (three isolates), Lb. kefiri (two isolates) and Lb. casei (1 isolate). The obtained results showed that the majority of colonies growing in the culture medium were Lb. plantarum. The probiotic properties of these isolates were found to maintain their viability in media containing low levels of acid and bile salt. Dogan and Ozpinar (2017) described that Lb. plantarum isolates exhibited probiotic properties. Leite et al. (2012) previously investigated the resistance of 34 lactic acid bacteria isolated from four different kefir grains to low pH and bile salt conditions, reporting that Leu. mesenteroides (18 isolates), Lc. lactis subsp. lactis (eight isolates), Lc. lactis subsp. cremoris (three isolates) and Lb. paracasei (five isolates) met the probiotic properties with regards to these two tested criteria. A study conducted by Zheng et al. (2013) investigated the probiotic properties of Lb. acidophilus, Lb. plantarum and Lb. kefiri samples isolated from Tibetan kefir grain. All of the isolates were identified as being resistant to pH 3 and 0.3% bile salt. In the present study, the bacteria that grew in the M17 agar medium and that exhibited resistance to low pH and bile salts were identified as E. faecalis (one isolate), E. faecium (one isolate), E. durans (one isolate), Str. thermophilus (two isolates), Lc. lactis subsp. lactis (one isolate) and Lc. lactis subsp. cremoris (one isolate). Scientific studies have demonstrated that the variety of lactic acid bacteria isolated from kefir grain and beverages differs from country to country. Each country and region are considered to have a unique population, and the probiotic properties of the bacteria obtained from these samples may also vary depending on the species and even strains. The phylogenetic tree of 23 lactic acid bacteria, drawn using the neighbor-joining method, is shown in Figure 5.
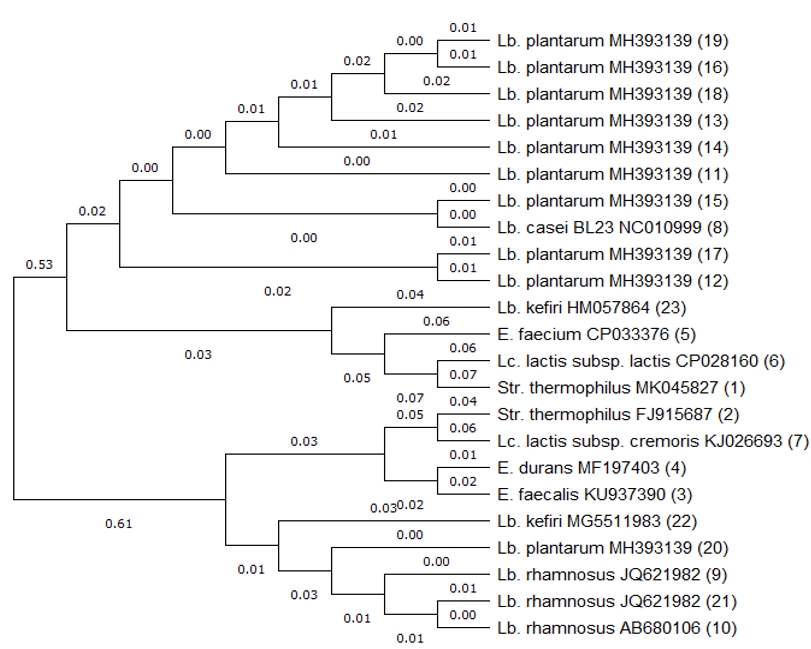
Figure 5. Phylogenetic tree based on 16S rRNA gene sequences of 23 isolates of lactic acid bacteria. Tree was constructed with neighbor-joining method. The numbers in parentheses indicate isolate numbers.
Conclusions
The content of the complex population obtained directly from the medium can be identified through a metagenomic analysis, although the DNA isolation method used and the quality of the obtained DNA are also important in such cases. While the results of the present study have revealed the greater microbial diversity of the homemade kefir samples, it was also shown that commercially available kefir beverages contain sufficient amounts of viable microorganisms, and that these microorganisms exhibit desirable probiotic properties such as acid, and bile salt resistance. It is therefore thought that further studies should be conducted to encourage the consumption of fermented milk products such as kefir and koumiss.
Acknowledgments
This study is a summary of the Ph.D. Thesis of the first author and is supported by the Academic Staff Training Program with the Project number of 2014-OYP-033. A part of this study was presented as an abstract in the 1st International Congress on Medical Sciences and Biotechnology, 14-16 October 2019 Uşak/Turkey.
Analiza mikrobiote kefira i kumisa s naglaskom na određena funkcionalna svojstva odabranih sojeva bakterija mliječne kiseline
Sažetak
Cilj ovog rada bio je odrediti mikrobiotu komercijalno dostupnog kefira, kumisa i kefira proizvedenog u kućnoj radinosti primjenom metagenomičke analize te usporediti neka probiotička svojstva bakterija mliječne kiseline izoliranih iz tih napitaka sa sojem Lactobacillus casei koji se koristi u proizvodnji yakulta. Kao uzorci su korišteni jedan kefir iz kućne radinosti, pet komercijalno dostupnih kefira različitih proizvođača i jedan kumis. Mikrobna raznolikost kefira i kumisa određena je metagenomičkom analizom na ciljano područje V1-V2 16S rRNK gena. Direktnom izolacijom DNK utvrđeno je da su u komercijalno dostupnim uzorcima kefira dominantne vrste Streptococcus thermophilus i Lactococcus lactis. Izolacijom na MRS agaru dominantnim su se pokazali Lc. lactis i Leuconostoc mesenteroides, dok je izolacijom na M-17 agaru Lc. lactis bila dominantna vrsta. U kefiru iz kućne radinosti koji je proizveden pomoću kefirnih zrnaca, dominantna vrsta je bio Lb. kefiranofaciens. Lb. kefiri i Enterococcus durans su se pokazali dominantnim vrstama izolacijom na MRS I M-17 agaru. Lb. kefiranofaciens, Lb. kefiri, i Str. thermophilus su bile dominantne vrste u kumisu. Mikroorganizmi izolirani iz kumisa pokazali su osnovna probiotička svojstva slično bakterijama mliječne kiseline izoliranim iz yakulta. Ovo istraživanje daje prikaz bakterijske mikroflore i probiotičkih svojstava bakterija mliječne kiseline izoliranih iz kefira i kumisa koji se konzumiraju u Turskoj.
Ključne riječi: kefir; kumis; bakterije mliječne kiseline; metagenomička analiza
References
https://doi.org/10.1016/j.lwt.2017.01.041
