INTRODUCTION
Heart failure (HF) is a syndrome characterized by a reduction in cardiac output due to primary cardiac injury, resulting in the inability to meet the metabolic needs of tissues despite normal venous return (1). Clinically, it is typically associated with systemic circulatory congestion of various etiologies and represents the advanced stage of cardiac pathology (2). Optimal intake of food, such as whole grains, fruits, nuts, dairy products, vegetables, legumes, eggs, fish, meats, and tea significantly reduces the risk of coronary heart disease, stroke, and high blood pressure, suggesting that the consumption of these foods in daily life is beneficial in reducing the risk of cardiovascular disease (CVD) (3). Green tea (Camellia sinensis) has become the most widely consumed beverage globally and plays a crucial role in regulating metabolism and cardiovascular health (4, 5). It effectively prevents detrimental cardiovascular events (atherosclerosis, hypertension, cardiomyopathy, ischemic heart disease, cardiac hypertrophy, and heart failure) and reduces cardiovascular mortality by decreasing oxidative stress, preventing inflammatory responses, and reducing platelet aggregation (6).
Lu'an GuaPian tea is a type of green tea cultivated in Lu'an City, Anhui Province, which holds a prestigious position as one of the top ten famous teas in China (7). The flavonoids present in Lu'an GuaPian tea primarily consist of flavonoid aglycones and flavonoid glycosides, with kaempferol-3-O-rutinoside (KR) being the most abundant among them (8). KR exhibits cardioprotective, antiatherosclerotic, antidiabetic, cerebral protective, hepatoprotective, anticancer, renoprotective, and antiadipogenic effects (9). Our previous study found that KR could inhibit inflammatory response through TLR4/MyD88/NF-κB signaling pathway in LPS-induced H9c2 cell injury (10). KR improved cardiac function, alleviated pathological changes in the heart, reduced excessive deposition of interstitial collagen in the myocardium, and inhibited cardiomyocyte apoptosis in rats with acute myocardial infarction (AMI). The protective effect of KR was associated with the regulation of the NF-κB/NLRP3/Caspase-1 signaling pathway (11). Our further study has revealed that KR has the potential to decrease the rate of apoptosis, reverse the elevated levels of IL-1β, IL-6, and TNF-α, and regulate the AMPK/SIRT1 signaling pathway in vitro (12). These results suggested that the cardioprotective effects of KR were associated with inhibition of the TLR4/MyD88/NF-κB, blockade of NF-κB/NLRP3/Caspase-1, and activation of the AMPK/SIRT1 signaling pathways. However, a comprehensive study of the mechanism of cardioprotective effects of KR is still unclear.
In this study, network pharmacology and molecular docking were employed to elucidate the mechanism of KR's cardioprotective effects. Subsequently, the predicted targets of KR against HF were validated through in vivo experiments to clarify its mechanism.
EXPERIMENTAL
Network pharmacology
Acquisition of KR targets. – The SMILES ID for KR was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) (13). The SMILES ID was then imported into the Swiss Target Prediction database (http://www.swisstargetprediction.ch/) (14) and the SEA database (http://sea.bkslab.org/) (15) to identify the targets of KR. The targets of KR were also retrieved through the PharmMapper database (http://www.lilab-ecust.cn/pharmmapper/) (16), the Batman-TCM database (http://bionet.ncpsb.org.cn/batman-tcm/) (17), and the GeneCards database (https://www.genecards.org/) (18).
Acquisition of disease targets. – The term "heart failure" was utilized as a keyword to conduct a search for human genes in the NCBI database (https://www.ncbi.nlm.nih.gov/) (19), GeneCards database (https://www.genecards.org/), and DisGeNET database (https://www.disgenet.org/) (20).
Acquisition of Venn diagrams. – The common target genes of KR and HF were acquired by Venny (version 2.1, https://bioinfogp.cnb.csic.es/ tools/venny/).
Construction of protein-protein interaction (PPI) network. – The common target genes of KR and HF were inputted into the String database (https://string-db.org/cgi/input.pl) to construct a PPI network (21). The protein type was specified as "Homo sapiens" and the PPI network data were retrieved. The results were visualized using Cytoscape 3.8.0 software, with node colors and sizes adjusted based on their degree values. Nodes were scaled larger and colored redder according to higher degree values.
GO enrichment analysis. – GO enrichment analysis comprises the assessment of biological processes (BP), cellular components (CC), and molecular functions (MF) with a corrected p-value < 0.05. The software utilized for drawing the histogram and bubble diagram includes R software (version R 4.1.2), clusterProfiler, enrichplot, and ggplot2.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. – The common targets of drug-disease were analyzed for KEGG pathway enrichment with a corrected p < 0.05. R software (version R 4.1.2) and clusterProfiler were utilized to generate the histogram and bubble diagram.
Molecular docking
CB-dock website was used to predict the binding affinity of KR and PI3K or AKT (1) phosphoinositide 3-kinase (PI3K, PDB ID: 1E8Y) (22) or AKT (PDB ID: 3D0E) (23) was downloaded from the RCSB PDB database. Water molecules and heteroatoms were deleted from targets (2). The structure of KR was downloaded from PubChem website (3). PI3K or AKT and KR were input to CB-Dock for molecular docking (24).
In vivo experiment
Animals. – A total of 24 Sprague-Dawley rats, male (200 ± 20 g), with manufacturing license SCXK 2017-001, and the experiment was approved by the Experimental Animal Ethics Committee of Anhui University of Chinese Medicine (AHUCM-rats-2023012).
Drugs and reagents. – KR was from Shanghai Yuanye Bio-Technology Company (China). Trizol (G3013) and SYBR (G3326-15) were from Servicevbio Biotechnology Co. Ltd. (China). TNF-α (MM-0180R1) and IL-6 (MM-0190R1) were purchased from Meimian (China). Anti-PI3K (WL02240) was purchased from WANLEIBIO. Anti-p-PI3K (R341468), anti-Akt (R23412), and anti-p-Akt (R22961) were from Abmart. Anti-GAPDH (BM3874) was purchased from BOSTER (USA).
Animal modeling and drug treatment
As mentioned previously, the model of AMI in rats was induced by ligating the left anterior descending (LAD) coronary artery (25). Two weeks later, the AMI rats were randomly assigned to three groups: model group (received normal saline via intragastric administration, 6 rats), KR group (received 10 mg kg–1 via intragastric administration, 6 rats), and captopril group (received 4.375 mg kg–1 via intragastric administration, 6 rats) for a duration of 4 weeks. Rats in the sham group (6 rats) and the model group received the same volume of normal saline intragastrically.
Hematoxylin-eosin (HE) staining. – The region of the left ventricular myocardium affected by myocardial infarction was collected, fixed with 4 % paraformaldehyde, and processed into thick tissue slices for paraffin embedding. HE staining was conducted to observe the pathological changes in the tissue.
Enzyme-linked immunosorbent assays (ELISA) for TNF-α and IL-6. – The myocardial tissue was homogenized with an appropriate amount of saline, and the supernatant was collected by centrifugation at 3000 rpm for 10 min. The levels of TNF-α and IL-6 in rat myocardial tissue were measured following the instructions provided with the ELISA kit.
RT-PCR for mRNA expression. – Total RNA was extracted from rat myocardial tissues of each group according to the instructions of the RNA Trizol kit, and cDNA was synthesized by reverse transcription after measuring the concentration, then PCR amplification was performed according to the instructions of SYBR kit, and the mRNA level of the target gene was calculated by the 2–ΔΔCq method. The primer sequence for PI3K (Forward: 5′-GCCTCAGTGGACTTGGATGTGTTC-3′, Reverse: 5′-GTCTTCGGAGCTTGGTACTTCTTGG-3′), AKT (Forward: 5′-GCCTCAGTGGACTTGGATGTGTTC-3′, Reverse: 5′-GTCTTCGGAGCTTGGTACTTCTTGG-3′), and β-actin (Forward: 5′-CCCATCTATGAGGGTTACGC-3′, Reverse: 5′-TTTAATGTCACGCACGATTTC-3′) were used in the study.
Western blot for protein expression. – Rat myocardial tissue was utilized for total protein extraction using RIPA lysate, and the BCA method was employed to determine the total protein concentration. The lysates were then separated on a 10 % to 15 % polyacrylamide gel and subsequently transferred to a PVDF membrane. Next, the PVDF membrane was blocked with 5 % skim milk before incubating with PI3K (1:1000), p-PI3K (1:1000), AKT (1:1000), p-AKT (1:1000), and GAPDH (1:5000) at 4 °C overnight. The secondary antibody was subsequently incubated with the primary antibody for 2 h at room temperature, followed by an overnight incubation at 4 °C. The ECL chemical substrate luminescence kit was utilized to measure the protein band density, and the Tanon 5200 imaging system (Tanon, China) was employed to capture images of the protein bands. The optical density values of the bands were analyzed using ImageJ software.
Statistical analysis
The data were analyzed using GraphPad Prism 9.4.1 software and expressed as mean ± standard deviation. Comparisons between multiple groups were conducted using ANOVA, with a significance level of p < 0.05 considered to indicate statistical significance.
RESULTS AND DISCUSSION
Network pharmacology
Integration of KR and HF targets. – 290 potential targets of KR were obtained, along with 3212 genes associated with HF. The screened KR and HF targets were input into the Venny 2.1 software, resulting in the identification of 151 common targets (Fig. 1).

Fig. 1. The Venn diagram of the common targets. The 151 common targets for KR in the treatment of HF.
PPI network and core target analysis
The nodes in the network represent the target genes, with the color depth and node size positively correlating with the degree value. The network consists of 151 nodes and 1639 edges, with an average degree value of 21.9. Notably, the top 10 core targets were ALB, TNF, AKT1, EGFR, CASP3, PTGS2, HSP90AA1, PPARG, ESR1, and GSK3B (Fig. 2).
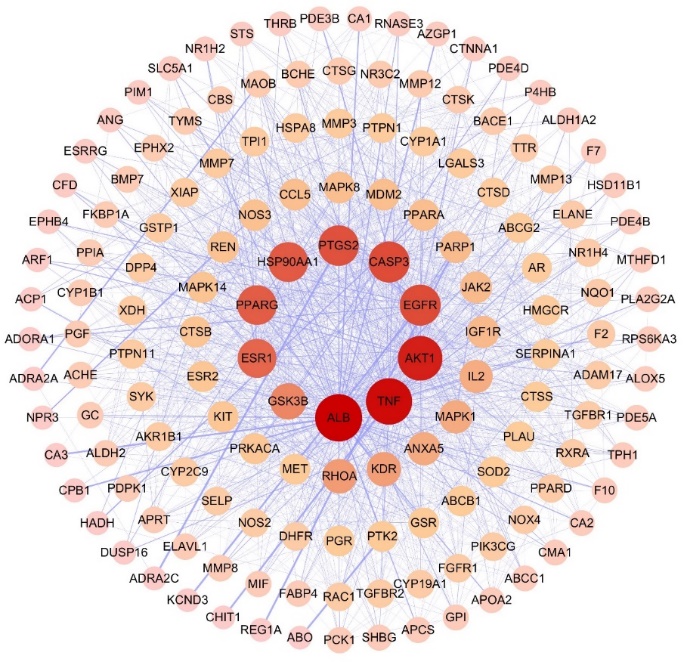
Fig. 2. PPI network diagram. The graph shows the interrelationships among 151 common targets.
GO enrichment result. – GO enrichment analysis revealed 1707 BP terms, 145 CC terms, and 53 MF terms. The top 10 entries were displayed based on the number of enriched genes (Fig. 3).

Fig. 3. GO enrichment analysis: BP, CC, and MF diagram.
KEGG enrichment result. – A total of 142 enrichment pathways were screened (Fig. 4). The results suggested that KR can be used to treat HF through lipid and atherosclerosis, PI3K-AKT signaling pathway, and MAPK signaling pathway. The PI3K-AKT signaling pathway may be the main mechanism, therefore, we chose this pathway for in-depth study.

Fig. 4. KEGG enrichment analysis. The horizontal axis represents importance according to the corrected p-value.
Molecular docking results
The results showed that the VINA docking score of KR with PI3K or AKT was higher, indicating that KR could bind well to both targets (Table I and Fig. 5). Molecular docking was validated against key targets of signaling pathways enriched by network pharmacology, followed by in vivo animal experiment validation.
Table I. Docking of KR with PI3K or AKT
| Targets | Vina score | Cavity score | Center (x, y, z) | Size (x, y, z) |
|---|---|---|---|---|
| PI3K | –9.7 | 9572 | 30, 0, 16 | 35, 24, 32 |
| AKT | –8.0 | 6227 | 26, –44, –2 | 31, 24, 32 |

Fig. 5. Pictures of KR docked to PI3K or AKT: a) KR-PI3K complex; b) KR binds to PI3K amino acid residues; c) KR-AKT complex; d) KR binds to AKT amino acid residues.
In vivo results
KR improved the morphological changes. – The myocardial tissue was orderly arranged, the cell structure was intact, the boundary was clear, the cytoplasm staining was uniform, and the nucleus was normal in the sham group. In the model group, the myocardial tissue boundary was unclear, the nucleus was fragmented, the cytoplasm was broken, the myocardial fibers were disordered, and the myocardial fibers were broken. The myocardial fibers were disordered and broken, with unclear texture direction, and accompanied by inflammatory cell infiltration in the myocardial interstitium. However, KR and captopril could reduce the fibroblasts, and alleviate the inflammatory cell infiltration, and most of the myocardial fibers were arranged neatly, with a tendency towards normal cell arrangement and relatively clear structure (Fig. 6).

Fig. 6. KR improved the morphological changes (×400): a) Sham group; b) model group; c) KR group; d) captopril group.
KR reduced the overproduction of inflammatory factors. – The results indicated a significant increase in the levels of TNF-α and IL-6 in the model group (p < 0.01), while their expressions were notably suppressed in the KR or Captopril groups (p < 0.01) (Fig. 7). There was no significant difference between KR and captopril on inflammatory factors.

Fig. 7. KR reduced the overproduction of inflammatory factors in AMI rats: a) TNF-α; b) IL-6. Compared with the sham group, **p < 0.01, compared with the model group, ##p < 0.01.
KR elevated the mRNA expressions of PI3K and AKT. – As depicted in Fig. 8, the mRNA expressions of PI3K and AKT were significantly decreased in the model group (p < 0.01). However, treatment with KR effectively reversed the decreased mRNA expressions of PI3K and AKT (p < 0.01). There was no significant difference between KR and captopril on the mRNA expressions of PI3K and AKT.

Fig. 8. KR elevated the mRNA expressions of PI3K and AKT: a) PI3K mRNA; b) AKT mRNA. Compared with the sham group, **p < 0.01, compared with the model group, ##p < 0.01.
KR elevated the protein expressions of PI3K and AKT. – Compared to the sham group, there was a significant decrease in the protein expression of PI3K and AKT in the model group (p < 0.05, p < 0.01). After administration of KR or captopril, there was a significant increase in the protein expression of PI3K and AKT (p < 0.05, p < 0.01) (Fig. 9). The results suggested that KR exerted cardioprotective effects by a mechanism related to activation of PI3K and AKT. There was no significant difference between KR and captopril on the protein expressions of PI3K and AKT.

Fig. 9. KR elevated the protein expressions of PI3K and AKT: a) p-PI3K/PI3K; b) p-AKT/AKT. Compared with sham group, *p < 0.05, **p < 0.01. Compared with the model group, #p < 0.05, ##p < 0.01.
Discussion
AMI is the result of myocardial necrosis caused by acute and persistent ischemia and hypoxia of the coronary arteries (25). Following AMI, myocardial tissues experience severe and sustained ischemia and hypoxia, leading to cardiac systolic and diastolic dysfunction as well as HF (26). AMI is one of the most common and important causes of HF and is associated with its resulting cardiomyocyte death. As the end stage of most CVDs, the long-term prognosis of HF after AMI is poor and imposes a significant disease burden on society (27). Therefore, searching for ideal drugs, especially medicinal foods to be consumed in daily life, can prevent the development of CVDs.
Network pharmacology is based on the theory of systems information biology and involves constructing a "drug-target-disease" visualization model to predict the action of drugs and disease targets. We used different databases to identify ingredient targets and disease targets (28). BATMAN-TCM is an online bioinformatics analysis tool designed for studying the molecular mechanisms of Chinese medicine ingredients, primarily used for predicting natural active ingredient targets (17). The Pharmmapper website uses a ligand-protein reverse docking strategy to explore potential ligand-binding sites (16). After conducting GO and KEGG enrichment analysis, we identified the potential signaling pathways through which KR may improve HF as lipid and atherosclerosis, PI3K-AKT signaling pathway, and MAPK signaling pathway. It is suggested that the PI3K-AKT signaling pathway may serve as the primary mechanism.
Molecular docking was used for the initial validation of KR targets, which refers to the application of computer technology for simulating the binding of ligands (such as proteins, DNA/RNA, and small molecules) and receptors (protein biomolecules), followed by the calculation of physicochemical parameters to predict binding mode and affinity. Essentially, it involves a process of mutual recognition between two or more molecules, encompassing spatial and energy matching (29, 30). Molecular docking involves the binding of natural active ingredients to specific target proteins and can elucidate the mechanism of natural active ingredients at the molecular level. This process allows for a detailed understanding of how these ingredients interact with their target proteins and provides valuable insights into their biological activity. In this study, KR showed that better binding affinity for PI3K and AKT, indicating a stronger binding capacity and validating the signaling pathway.
We used in vivo experiments to verify the relevance of the mechanism of KR against HF to the regulation of the PI3K/AKT signaling pathway, which plays an important role in regulating numerous physiological processes through activating downstream effectors involved in cell cycle transitions and cell proliferation (31). Activation of the PI3K/AKT signaling serves a protective role in compensatory hypertrophic responses and contractile dysfunction and even prevents cardiomyocyte apoptosis (32). Decreased PI3K/AKT phosphorylation is a characteristic feature in the pathogenesis of AMI. The activation of PI3K/AKT phosphorylation has been shown to effectively alleviate AMI and myocardial reperfusion injury (33). In HF, cardiac tissues are damaged, and PI3K activates AKT by phosphorylation to produce effects, and the PI3K/AKT signaling pathway plays an important role in cardiomyocyte growth and repair (34). PI3K is an important kinase that catalyzes the conversion of inositol and phosphatidylinositol in eukaryotic organisms. P-AKT, a serine/threonine protein kinase with a highly conserved structure and function, is a key target downstream of PI3K and is widely expressed in human cardiac, brain, and lung tissues (35). PI3K is a dimeric protein composed of two parts, a catalytic subunit (p110) and a regulatory subunit (p85). When receiving a stimulus signal, PI3K is activated by converting the substrate phosphatidyl inositol diphosphate (PIP2) into phosphatidyl inositol triphosphate (PIP3), which further binds to AKT, causing AKT to be recruited to the membrane and activated by phosphorylation. The activated PIP3 further binds to AKT, which is then recruited to the membrane and activated by phosphorylation. NF-κB is downstream of the PI3K/AKT signaling pathway and acts as a transcription factor that is activated by AKT phosphorylation and then translocated to the nucleus, regulating the encoding and translation of specific nucleic acid sequences, releasing TNF-α and IL-6, and activating inflammatory responses (36, 37). In this study, KR improved the morphological changes, reduced the levels of TNF-α and IL-6, and enhanced the mRNA and protein expressions of PI3K and AKT, which indicated that KR ameliorated HF after AMI through enhancing the expressions of PI3K and AKT. There was no significant difference between KR and captopril on the levels of TNF-α and IL-6, mRNA, and protein expressions of PI3K and AKT. Thus, the effect of KR is similar to that of captopril, KR can be obtained from food as a natural product and can be ingested as a homologous product of medicine and food.
CONCLUSIONS
Network pharmacology and molecular docking were utilized to predict the targets of KR and its associated signaling pathway as the PI3K/AKT pathway. Our findings suggest that KR may be a viable therapeutic option for protecting against HF. The application of KR in the treatment of HF presents a feasible strategy for reducing AMI and improving cardiac function. These results offer promising novel approaches to enhance the management and prognostic strategies for HF.
Data availability. – All datasets are available from the corresponding author upon reasonable request
Conflicts of interest. – The authors declare no conflict of interest.
Funding. – These works were supported by the Key Project Foundation of Natural Science Research in Universities of Anhui Province in China (2022AH050479), the Outstanding Youth Scientific Research Project of Anhui Universities (2022AH030158), and the Teaching Team Project of Anhui Xinhua University (2022jxtdx03).
Authors contributions. – Conceptualization, F.H. and P.Z.; methodology, L.Q.G.; analysis, L.Q.G. and L.Z.; investigation, L.Q.G., L.Z., S.N.L., J.B. and L.L.S.; writing, original draft preparation, L.Q.G.; writing, review and editing, F.H. and P.Z. All authors have read and agreed to the published version of the manuscript.
