Introduction
Rivaroxaban, a factor Xa inhibitor, is the most prescribed of the non‐vitamin K antagonist oral anticoagulants (NOACs), a class of drugs that is progressively replacing classical oral anticoagulants (vitamin K antagonists, VKA) worldwide, in a wide range of indications (1-3). As is the case with other NOACs (Xa inhibitors apixaban and edoxaban, and a direct thrombin inhibitor dabigatran), rivaroxaban is favored over VKAs for simplicity of use that does not require constant monitoring of the coagulation cascade and, generally, for a lower risk of major bleedings (4). However, exposure to and anticoagulant effect of rivaroxaban is affected by age, sex, body weight, hepatic and renal function, concomitant diseases and treatments (5,6).
Interactions between rivaroxaban and other drugs are typically pharmacokinetic and are based on the fact that rivaroxaban is a substrate of cytochrome P-450 enzymes CYP3A4/5 and CYP2J2 (and several CYP-independent mechanisms), as well as of two major efflux transporter proteins – ABCB1 (multidrug resistance protein 1, MDR1 or P-glycoprotein, P-gp) and ABCG2 (breast cancer resistance protein, BCRP) (7,8). There is some evidence that polymorphisms in genes encoding the respective metabolizing enzymes and/or transporter proteins (CYP3A4, CYP3A5, CYP2J2, ABCB1, ABCG2) might affect exposure to and efficacy/safety of rivaroxaban, but at present, the results are equivocal and insufficient for implementation in clinical practice (9-11).
ABCB1 protein is a crucial efflux membrane transporter with a protective function, and with a wide range of endogenous and exogenous substrates, including many drugs. Located at the apical membrane, it excretes orally administered drugs back into the small intestine and colon, in the kidney it excretes substances into the tubular lumen, and in the liver it excretes them into the bile, thus reducing their circulating levels (12-14). Although there is evidence for interindividual differences in the ABCB1 expression and transport function, the genetic contribution is still not fully understood (15-17). It has been implied that ABCB1 rs1128503, rs2032582 and rs1045642 polymorphisms reduce the ABCB1 transport function in vitro, but that the effect might be substrate-dependent (18). The fundamental in vitro research concerning the association of rivaroxaban with the ABCB1 transport function was published 10 years ago – cellular efflux of rivaroxaban was markedly reduced by two ABCB1 inhibitors, whereas its clearance was greatly reduced in knock-out mice lacking ABCB1 (and ABCG2) transporters (Mdr1a/Mdr1b(-/-)/Bcrp(-/-) mice) (19,20). The ABCB1 gene is highly polymorphic. The most common (particularly in European population) and the most extensively investigated ABCB1 polymorphisms are three coding polymorphisms: rs1128503 (ABCB1 c.1236C>T); rs2032582 (ABCB1 c.2677G>T/A) and rs1045642 (ABCB1 c.3435C>T) (21). These polymorphisms are in a strong linkage disequilibrium (LD), and haplotypes with variant alleles (e.g., TTT) – compared to wild type (CGC) – are associated with altered ABCB1 protein folding, binding site conformation and reduced transporter activity in vitro (11,22,23). Among other ABCB1 polymorphism, rs4148738 (ABCB1 c.2482-2236G>A) is relatively common in European population (21). This intronic polymorphism was also found to affect the pharmacokinetics of ABCB1 substrates (11). The first indication that the ABCB1 c.3435C>T (rs104566642) and c.2677G>T (rs2032582) polymorphisms might be associated with the bleeding risk in rivaroxaban-treated patients was based on a case report (24). Therefore, it is plausible to assume that these four common ABCB1 polymorphisms – through the resulting reduced transporter activity – might increase bioavailability of rivaroxaban. This, in turn, could reflect on its anticoagulant activity. Consequently, we aimed to estimate whether they affected the risk of bleeding in rivaroxaban-treated patients.
Materials and methods
Subjects and study design
Presented data are part of a larger prospective nested case-control study (“Pharmacogenomics in Prediction of Cardiovascular Drugs Adverse Reaction”) that started December 15, 2020 and will last 60 months and include 1200 subjects. The study (ClinicalTrials.gov, NCT05307718) is conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committees of the University of Zagreb, School of Medicine (reg. number 380-59-10106-20-111/125; class 641-01/20-02/01) and the University Hospital Centre Zagreb (class 8.1-20/142-2; number 02/21 AG), Zagreb, Croatia. The primary cohort includes adults (> 18 year of age) with a new-onset indication for treatment with NOACs, antiplatelets and/or statins. All subjects provided written informed consent.
The present analysis included consecutive consenting adults (≥ 18 years of age) who started rivaroxaban for any indication requiring > 6 months of therapy, and were followed-up until occurrence of bleeding, or if no bleeding occurred, for > 6 up to 12 months within the time period between December 15, 2020 and March 1, 2023.
Comedication present at baseline or in place for at least one month before the bleeding event in cases, or in place for at least (any) 3 months in controls, was classified as (enzyme or transporter) substrates, inducers or inhibitors using the Lexicomp Clinical Decision Support System (25).
Patients who developed major or clinically relevant non-major bleeding, as defined by the International Society on Thrombosis and Haemostasis (ISTH), within the first 6 months of treatment were considered cases, whereas controls were patients who experienced no bleeding over > 6 months of treatment (26,27).
Major bleeding is defined as fatal bleeding, and/or symptomatic bleeding in a crucial area or organ, and/or bleeding resulting with a decrease in hemoglobin concentrations of ≥ 20 g/L, or indicating a transfusion of ≥ 2 units of whole blood or red cells (26). Non-major bleeding is defined as multiple-source bleeding, unexpected hematoma (> 25 cm2), epistaxis (> 5 minutes), gingival bleeding (> 5 minutes), macroscopic hematuria, rectal bleeding, coughing or vomiting blood, vaginal bleeding, blood in semen, intra-articular bleeding with trauma, or surgical-site bleeding (27).
All present subjects were recruited at a single tertiary centre. They were treated and followed-up in line with the standard of care by physicians specialized in management of the respective conditions (e.g., non-valvular atrial fibrillation, deep vein thrombosis, pulmonary embolism, secondary prophylaxis after acute coronary syndromes, primary prophylaxis in patients with coronary artery disease or peripheral artery disease). Patients were instructed to contact their prescribing physician regardless of their regular scheduled visits in case of any bleeding, which was then assessed in line with the ISTH criteria (26,27). Attending physicians were not aware of the patients’ pharmacogenetic status at the time of assessment of severity of bleeding or confirmation of the “control” status. The “control” status for patients not experiencing bleeding over > 6 months of treatment was verified by telephone contacts at the cut-off date defined for the purpose of the present analysis.
Blood sampling
At the inception of the cohort, blood samples were taken for the genetic analysis (3 mL of whole blood) in an K3EDTA tube (Vacuette, Greiner Bio-One International AG, Kremsmünster, Austria). For routine biochemical, hematological and coagulation analyses following blood samples were taken: 8 mL of serum in a test tube without biochemical additives (Vacuette, Greiner Bio-One International AG, Kremsmünster, Austria) and/or 3 mL of whole blood in a K3EDTA tube (Vacuette, Greiner Bio-One International AG, Kremsmünster, Austria) and/or 2.7 mL of plasma in a sodium citrate tube 0.105 M (3.2%) (Becton Dickinson, Plymouth, United Kingdom), respectively. Routine biochemical, hematological and coagulation analyses were performed according to the attending physicians’ orders.
Isolation of DNA and pharmacogenetic analyses
Genomic DNA was extracted from whole blood using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Pharmacogenetic analyses were performed by using specific TaqMan DME and SNP Assays on 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, USA) for genotyping of CYP2J2, CYP3A4, CYP3A5, ABCB1, and ABCG2 gene variants (28). Only for the ABCB1 triallelic locus c.2677G>T/A (rs2032582) genotyping was performed by the real-time PCR on the LightCycler v 2.0 device (Roche Diagnostics, Germany), as described by von Arjomand-Nahad et al. (29).
We investigated four ABCB1 polymorphisms of primary interest: rs1128503 (assay ID C___7586662_10), rs2032582, rs1045642 (assay ID 4362691 C___7586657_20) and rs4148738 (assay ID C___1253813_10). We determined additional gene polymorphisms with possible effect on rivaroxaban pharmacokinetics, and considered them as covariates (confounders): the common loss-of-function ABCG2 polymorphism c.421C>A, rs2231142 (assay ID 4362691 C__15854163_70), CYP3A4 *1B, rs2740574 (assay ID 4362691 C___1837671_50), and *22; rs35599367 (assay ID 4351379 C__59013445_10) and CYP3A5 *3, rs776746 (assay ID C__26201809_30) polymorphisms needed to determine CYP3A4/5 genotype-predicted phenotype, and CYP2J2 A>T (rs11572325, assay ID C__30760106_10) and *7 (rs890293, assay ID 4362691 C___9581699_80) polymorphisms to determine CYP2J2 phenotype. CYP3A4/5 genotype-predicted phenotype is defined as follows: extensive metabolizer – high activity (CYP3A4*1/*1 and CYP3A5*1 carriers); intermediate metabolizer – intermediate activity (CYP3A4*1/*1 or CYP3A4*22 carriers and CYP3A5*3/*3 or CYP3A5*1 carriers) and poor metabolizer – low activity (CYP3A4*22 carriers and CYP3A5*3/*3) (30). CYP2J2 genotype-predicted phenotype is defined as high activity (CYP2J2 *1/*1) and intermediate or low activity (CYP2J2 *7 carriers) (31,32).
Statistical analysis
We expected the four ABCB1 polymorphisms to be in a strong pairwise LD, and we planned to classify the subjects based on the “load” of variant alleles across them as: i) 0-2 variant alleles, but no locus is variant homozygous (i.e., wild-type or a maximum of 2 heterozygous loci); ii) 2-5 variant alleles, i.e, at least one variant homozygous locus to a maximum of 5 variant alleles (2 variant + 1 heterozygous or 1 variant + 3 heterozygous loci); iii) 6-8 variant alleles (17,20,25-28). We further expected that distribution of patients across these categories would be 30%, 50% and 20%, respectively, and that 15-20% would experience bleeding over the first 6 months of treatment (18,33). We approximated that with such prevalence of genotypes and events, a sample of 250-300 patients would provide 80-85% probability to detect a strong, clear increasing trend (the theoretical background suggests that variant alleles result in reduced ABCB1 function) in proportion of cases across the levels of ABCB1 variant alleles (e.g., 10%-20%-30%) (34).
We used energy balancing with average treatment effect as the estimand (package WeightIT in R Statistical Software, v4.1.2, R Core Team 2021) to achieve a balance between cases and controls regarding a number of demographic, comorbidity, (co)medication and pharmacogenetic variables (except for the ABCB1 polymorphisms) that could have confounded the relationship between the polymorphisms of interest and the case/control status (35,36). Energy balancing is a weighting method that achieves (where possible) a distributional balance of covariates between groups (37,38). Standardized differences (d) < 0.1 indicate an adequate balance, i.e., irrelevant differences. Sporadic covariates that could not be adequately balanced (d ≥ 0.1) were included in a multivariable weighted logistic regression model with robust variance estimation to generate estimated (adjusted) probabilities of bleeding. We report: a) raw and weighted (after energy balancing) proportions of patients with different “load” of ABCB1 variant alleles in cases and controls; b) raw, weighted (after energy balancing) and fully adjusted (from the logistic model) proportion of cases across the patient subsets with different “load” of ABCB1 variant alleles, and the associated test for trend in proportions. To supplement this analysis, we report also prevalence of all identified halplotypes and genotypes, and raw proportions of “cases” (incidence of bleeding) across them. We used SAS for Windows 9.4 (SAS Inc., Cary, USA). We used webtool CubeX (http://apps.biocompute.org.uk/cubex) to determine Hardy-Weinberg equilibrium and LD (39).
Results
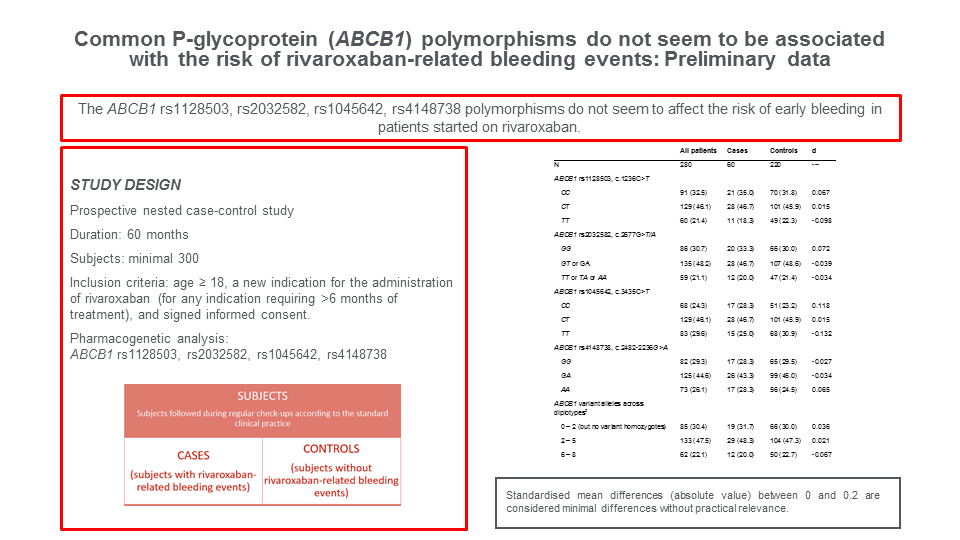
There were a total of 60 cases and 220 control patients (Table 1). Cases were older (mean 73 vs. 63 years, d = 0.799) and had somewhat lower estimated glomerular filtration rate (eGFR) (mean 65 vs. 74 mL/min/1.73 m2, d = - 0.372). Three of the case patients experienced a major bleeding (2 intracranial, 1 hemopericardium), while the rest were non-major bleedings, predominantly gastrointestinal. Rivaroxaban doses and co-treatment with antiplatelets were fairly similar in cases and controls (all d < 0.300). Cases and controls somewhat differed in prevalence of major comorbidities, with a clear difference in prevalence of malignant diseases (35% vs. 15.9%, d = 0.449) and gastrointestinal diseases (typically peptic acid disease and/or diverticulosis; 46.7% vs. 6.8%, d = 1.008) (Table 1).
Prevalence of genotypes across the four ABCB1 polymorphisms was closely similar in cases and controls (all d < 0.2) (Table 2). In line with the expectations, polymorphisms were in a strong pairwise LD (rs1128593 vs. rs2032582 D’ = 0.878, r2 = 0.750, Chi2 = 209.9; rs1128593 vs. rs1045642 D’ = 0.890, r2 = 0.574, Chi2 = 164.2; rs1128593 vs. rs4148738 D’ = 0.892, r2 = 0.680, Chi2 = 190.4; rs2032582 vs. rs1045642 D’ = 0.876, r2 = 0.561, Chi2 = 157.1; rs2032582 vs. rs4148738 D’ = 0.725, r2 = 0.331, Chi2 = 144.0; rs1045642 vs. rs4148738 D’ = 0.732, r2 = 0.469, Chi2 = 143.1), hence subjects were classified as planned, based on the “load” of variant alleles across the four loci as those with 0-2 variant alleles, but no variant homozygous locus (reference group), those with 2-5 variant alleles and those with 6-8 variant alleles: distribution of cases and controls across these three groups was closely similar (all d < 0.1). Prevalence of patients using different numbers of ABCB1 substrates or inductors was closely similar in cases and controls (all d < 0.1) (Table 2). Only the proportions of patients using 2 or ≥ 3 ABCB1 inhibitors were slightly different in cases than in controls (41.7% vs. 31.4%, d = 0.215, and 33.3% vs. 39.5%, d = -0.129, respectively) (Table 2).
Cases and controls were closely similar regarding the prevalence of ABCG2 c.421C>A variant carriers and ABCG2 substrate or inhibitor users (all d < 0.1) (Table 3); CYP3A4/5 polymorphisms and predicted phenotype, as well as the prevalence of CYP3A4/5 substrate users (all d < 0.2), with somewhat more CPY3A4/5 inhibitor users among controls (30.0%) than among (d = 0.232) cases (20.0%). They were also similar regarding prevalence of CYP2J2 genotypes and predicted phenotype, whereas less cases than controls were using CYP2J2 inhibitors (13.3% vs. 25.0%) (d = -0.300) (Table 3).
To estimate the association between the “load” of variant alleles across the four ABCB1 polymorphisms and the “case status” (bleeding), cases and controls were subjected to energy balancing regarding age, sex, eGFR, rivaroxaban dose, platelet co-treatment, relevant comorbidities, ABCG2 c.421 genotype, CYP3A4/5 and 2J2 phenotypes, and exposure to enzyme and transporter substrates and inhibitors (Table 4). Excellent balance was achieved (d < 0.1) for almost all covariates except that age (mean 70 vs. 67 years, d = 0.298) and proportions of patients with a history of cancer (26.4% vs. 21.8%, d = 0.109) and with gastrointestinal diseases (25.0% vs. 14.0%, d = 0.279) were still somewhat higher in cases then in controls (Table 4). Also, exposure to CYP2J2 inhibitors was less common among cases (18.3% vs. 22.7%, d = -0.172) (Table 4).
In this balanced (pseudo)population, prevalence of patients with 0-2, 2-5 or 6-8 variant alleles across the ABCB1 polymorphisms was similar in cases and controls: 30.5% vs. 30.8% (d = -0.007) for 0-2 variant alleles (no variant homozygous loci), 54.2% vs. 47.3% (d = 0.140) for 2-5 variant alleles, and 15.3% vs. 21.9% (d = -0.172) for 6-8 variant alleles (Table 4), just as was the case considering raw (unbalanced) data (Table 2). In reverse,Figure 1 shows proportions of cases across the patient subsets based on the increasing load of variant alleles across the four ABCB1 polymorphisms: raw proportions, weighted proportions based on balanced data and estimated probabilities from a multivariable logistic model for balanced data with additional adjustment for age, history of cancer, gastrointestinal diseases and use of CYP2J2 inhibitors – there is no obvious trend in the proportion of cases (bleeding) with increasing number of variant alleles.
We identified 15 out of 16 possible haplotypes. The completely variant (c.1236T_c.2677T(A)_ c.3435T_c.2482-2236A) and completely wild-type (C_G_C_G) haplotypes by far prevailed (40.7% and 39.5%, respectively), with closely similar prevalence of cases (patients who experienced bleeding) (21.1% vs. 23.1%, respectively). All other haplotypes were observed with frequencies from 0.2% to 6.4% (cumulatively 19.8%), with 18% prevalence of cases (not shown). We observed 34 genotype combinations across the four polymorphisms (c.1236C>T, c.2677G>T(A), c.3435C>T, c.2482-2236G>A), most common of which was one with 4 variant alleles (CT/GT(A)/CT/GA, N = 82, 29.3%), followed by the completely wild-type combination (CC/GG/CC/GG, N = 53 (18.9%) and the completely variant combination (TT/TT(A) /TT/AA, N = 49, 17.5%). Prevalence of cases was closely similar - 20.7% vs. 26.4% vs. 22.4%, respectively. All other genotype combinations were observed with frequencies between 0.3% to 6.8% (cumulative N = 96, 44.3%), with cumulative prevalence of cases of 18% (not shown).
Discussion
The purpose of pharmacogenomics research is to identify genetic traits that could be used as a prirori indicators of clinically relevant risks of failures or adverse effects of particular treatments in individual patients. With respect to this ultimate goal, the present analysis suggests that the four common ABCB1 polymorphisms (rs1128503, rs2032582, rs1045642, rs4148738) – by far the most prevalent of all ABCB1 polymorphisms in White Europeans – do not seem to have a clinically relevant effect on the risk of bleeding over the initial 6-12 months in the de novo rivaroxaban-treated patients. The study was conceived on the following grounds: (i) a reasonably expected incidence of bleeding of 20% over the first 6 months (33); (ii) the theoretical background suggesting that an increasing “load” of variant alleles across these polymorphisms could be reasonably expected to result in reduced ABCB1 function, ie., increased rivaroxaban bioavailability and anticoagulant effect; (iii) we considered that “clinical relevance” might be indicated by a robust trend of increased risk of bleeding, such that the rate of 20% is resultant to a low rate (e.g., 10%) with no or only a few variant alleles across the 4 polymorphisms, and then higher (e.g., 20%, 30%) with higher number of variant alleles (18). Under such circumstances, the fact that we observed consistently similar risks of bleeding and no indication of any trend in patients with different “variant loads” (raw data, fully adjusted analysis), similar risks with “fully wild-type” and “fully variant” haplotypes, and with “fully variant” and “fully wild-type” genotype combinations across the four polymorphisms, justifies a conclusion about the lack of signal of any “practically relevant” effect.
The reports on the ABCB1 polymorphisms and the risk of bleeding (major or clinically relevant non-major bleeding (CRNMB)) have thus far yielded controversial results. Comparison of outcomes in observational studies, particularly those involving genetic traits, is considerably less straightforward than in the case of randomized experiments because apparent differences or similarities could be due to many reasons other than the true underlying biological phenomenon that is actually evaluated, e.g., chance and sample size (small samples and/or low number of events are highly susceptible to chance findings), target population and the sample particulars, study design, ethnic and geographical determinants, control of confounding and susceptibility to other biases imminent in observational data. In a study based on administrative data, 999 Finnish patients newly started on rivaroxaban due to non-valvular atrial fibrillation (NVAF), vascular disease, pulmonary embolism (PE), deep venous thrombosis (DVT) or cerebrovascular incidents (CVI), were followed-up over a median period of 4 months (40). With adjustment for concomitant antiplatelet use, wild-type patients and variant carriers at each of the four ABCB1 polymorphisms (rs1128503, rs2032582, rs1045642, rs4148738) had a virtually identical risk of major or non-major clinically relevant bleeding, and the same was observed in the haplotype analysis (40). However, there were only 26 events – a few events more or less among either patient stratum (which could have occurred with essentially identical probability as the observed numbers), could have substantially changed the results (40). In a sample of 95 rivaroxaban-treated Chinese NVAF patients followed-up over one year, raw prevalence of genotypes at ABCB1 polymorphisms rs1128503, rs1045642 and rs4148738 was closely similar between those who experienced bleedings and those who did not – but there were only 16/95 patients with bleeding events (41). Obviously, data is extremely fragile. Likewise, a report that included 155 NVAF patients of Mongolian descent started on rivaroxaban, reported similar raw prevalence of genotypes at ABCB1 polymorphisms rs1128503, rs1045642 and rs4148738 in patients who experienced bleeding over the initial 7-10 days of treatment and in those who did not, but there were only 24 of the former, and the observational period was extremely short (42). In a cross-sectional study in 128 Russian NVAF patients older than 80 years of age with at least 7 days of treatment with rivaroxaban, 23 had a history of CRNMB (43). The authors report higher raw proportion of “bleeders” among rs1045642 variant homozygotes (12/41) vs. wild-type subjects (1/22), and among rs4148738 variant homozygotes (11/28) vs. wild-type subjects (3/37) – however, no control of confounding was undertaken, and the number of subjects and events (particularly among “wild-type” patients) was extremely low (43). Largely opposite results were reported in another study in Russian patients: 100 subjects with acute coronary syndrome and NVAF were treated with rivaroxaban and dual antiplatelet treatment for up to 12 months, and 38 developed major bleeding or CRNMB (44). The authors report an increased risk of bleeding as unadjusted (raw) odds ratio for rs1045642 wild-type subjects vs. variant carriers (exactly the opposite from the report in NVAF patients) of 3.13 (95%CI 1.03-9.52) (43). However, in addition to the fact of no confounding control, the authors erred in the calculation of the odds ratio – there were 13 “bleeders” among 29 wild-type subjects and 25 “bleeders” among 71 variant carriers, which gives OR = 1.50 (95%CI 0.62-3.58, P = 0.370). The authors also report unadjusted OR for rs4148738 variant carriers vs. wild-type subjects of 7.08 (95%CI 2.17 -23.1), but again err in the odds ratio calculation: there were 5 “bleeders” among 18 wild-type patients, and 33 “bleeders” among 82 variant carriers which gives OR = 1.75 (95%CI 0.57-5.38, P = 0.328) (we calculated Mantel-Haenszel ORs and CIs, and respective test statistics) (44).
Three further studies could be considered less informative on the topic, because they jointly considered patients treated with different NOACs. A single-centre registry analysis in the USA included 2364 White outpatients with NVAF followed-up over 1-3 years (45). With a comprehensive confounding control, the risk of major/CRNM bleeding was closely similar in wild-type patients and variant carriers at rs1128503, rs2032582, and rs1045642 considered individually and as haplotypes, but patients treated with rivaroxaban (N = 802) and apixaban (N = 1324) were considered jointly (45). Similarly, a cross-sectional case-control study in Korean patients (50 with bleeding and 418 controls) jointly considered patients on apixaban, edoxaban, dabigatran and rivaroxaban (N = 74), and suggested no univariate association between the case status and ABCB1 polymorphisms rs1128503, rs2032582 or rs1045642 (46). In contrast, another cross-sectional study in Korean patients (64 cases and 229 controls) treated with either apixaban or rivaroxaban (numbers not reported) suggested higher risk in variant carriers at rs1045642 vs. wild-type subjects (47). However, the reported OR (3.2, 1.35-7.43) was derived from a model with 10 covariates and was likely severely biased away from the null, since there were only 64 cases, and no measures were undertaken to reduce the bias (48).
The obvious limitation of the present study is a moderately-sized single-centre sample. However, from the purely “technical” standpoint, by using energy balancing to control for 18 of the 22 plausibly relevant confounders, we achieved a situation in which the final logistic model with 5 independents and effective sample of 58 cases and 205 controls enabled us to generate estimated probabilities of bleeding across the levels of ABCB1 variant allele “loads” reasonably protected from bias inherent to logistic models with a limited number of events. The confounders that we accounted for comprehensively addressed demographic, comorbidity, co-medication and genetic factors, and were observed and captured in real-time. As supported by the HWE tests, classification of patients regarding the ABCB1 and other polymorphisms was most likely correct, and the prospective study design ascertained adequate classification of “cases” and “controls”, particularly since all controls had at least 7 and a maximum of 10 months of follow-up without bleeding. We limited the “case” designation to patients who experienced bleeding within the initial 6 months, thus minimizing the interference of post-baseline (intercurrent) events that might have been difficult to control. The reasoning that if some genetic trait indeed has an important impact on the bleeding risk, this should be obvious already over the first 6 months of treatment is also medically justified: in the cited Finnish study, practically all bleeding events occurred within the initial 6 months (40).
Finally, our approach of categorization of patients with respect to their genotypes across the four strongly linked polymorphisms based on the “load” (number) of variant alleles might be objected. However, we find it to be biologically plausible – it reflects the underlying rationale of (presumably) altered ABCB1 transporter function consequent to the presence of variant alleles: subjects classified as those with 0 (wild type) to 2 variant alleles (out of 8 possible) but with no variant homozygosity could be reasonably viewed as those with no or minimal (hypothetical) alterations of the transporter functions, whereas those with 6 to 8 such alleles (all four loci variant homozygous) could be viewed as those with maximal (hypothetical) alterations. Even if variant alleles on different loci “favored” different (hypothetical) effects, it is the “net” effect of the entire set-up across the four polymorphisms that would be informative: an increasing trend in bleeding incidence across such a “variant gradient” would indicate their practical relevance. The fact of similar raw and weighted (adjusted) prevalence of genotypes at each of the four polymorphisms between cases and controls, and comparable probabilities of bleeding in the haplotype analysis and analysis of genotype combinations support the main observations.
In conclusion, the present nested case-control study strongly suggests that in White Europeans the common coding ABCB1 polymorphisms rs1128503 (c.1236C>T), rs2032582 (c.2677G>T/A), rs1045642 (c.3435C>T) and rs4148738 (2482-2236G>A) do not have any clinically relevant effect on the risk of bleeding over the initial 6 months in rivaroxaban-treated patients.