Introduction
Although many probiotic bacteria have been reported until now, new potential strains need to be studied (Binda et al., 2020; Marinova et al., 2019). Probiotic properties are specific to each strain; therefore, studies should be conducted to evaluate new probiotic candidates. Fermented dairy products are sources of probiotic bacteria. The microbial diversity of fermented foods is increased by conventional fermentation (Tamang et al., 2016). The health benefits of probiotics have been known for many years, and Lactobacillus, Lactococcus, Carnobacterium, Enterococcus, Streptococcus, Pediococcus, Vagococcus, Leuconostoc and Pediococcus are the best-known probiotic lactic acid bacteria (Pinto et al., 2020). Lactobacilli, which occur naturally in the gut microbiota, exhibit antimicrobial properties against many pathogens by producing organic acids, hydrogen peroxide (H 2O 2) and bacteriocins. It may also lower cholesterol, influence the immune system, and have antioxidant and antidiabetic properties (AlKalbani et al., 2019; Singhal et at., 2019). Probiotics with strong probiotic properties can produce healthy new functional probiotic foods.
Probiotic strains should resist low pH and bile salts and survive in the gastrointestinal tract environment (GIT). They should also multiply in the intestine. In addition, probiotic strains must be safe and maintain their viability in food throughout their shelf life (Binda et al., 2020). Probiotics should also have other functional properties. Probiotics with antioxidant properties can prevent many diseases related to oxidative stress and reduce the risk of breast and colon cancer (Liu et al., 2010; Amaretti et al., 2013). Probiotics with antimicrobial activity can be used as biopreservatives (Yong et al., 2015). Cheese is a very important component of the human diet (Saric et al., 2022). Tulum cheese has a distinctive flavour and is one of the most popular cheeses in Turkey (Albay and Şimşek, 2022). This study was conducted to determine the probiotic potential of Lactobacillaceae isolated from traditionally produced Tulum cheese from Turkey.
Materials and methods
Lactobacillaceae isolation
In this study, 32 individual isolates/colonies were randomly selected from MRS agar and purified three times by subculturing on the appropriate MRS medium. For the Lactobacillus isolates, cream-coloured and smooth-edged colonies were isolated (Kanak and Yılmaz, 2020). These colonies were isolated from fermented Tulum cheese sold in local markets in Sakarya province, Turkey. The Tulum cheeses were made from pasteurised cow's milk. man, Rogosa and Sharpe (MRS) (Merck, Germany) broth was added to 1 mL of each sample and incubated for 24 hours at 37 °C under anaerobic conditions. The enriched samples were diluted and plated out on MRS agar. This was incubated for 48 hours at 37 °C under anaerobic conditions. After incubation, each isolated colony was stored at -80 °C in MRS with 30 % glycerine.
Identification of bacteria with MALDI-TOF MS biotype
The MALDI-TOF MS (Matrix Supported Laser Desorption/Ionisation Flight Time Mass Spectrometry, Bruker, Germany) was used to identify the bacteria. A single colony was taken from the bacterial isolates developed on MRS agar and spread in a spiral on the target spot of the 96-well plate. 1 µL of 10 mL of the stock solution prepared with 70 % formic acid was dripped on and allowed to dry at room temperature. Then 1 µL of the matrix (α-cyano-4-hydroxycinnamic acid in 50 % acetonyl acid and 1.5 % trifluoroacetic acid) was dropped on the sample and dried again at room temperature. The prepared spots were loaded into the instrument for analysis. Samples were analysed automatically using a MALDI-TOF mass spectrometer (Bruker, Germany) and Flexcontrol 3.4 software. The analysis was performed for 10 strains identified as Lactobacillaceae.
Lactobacillaceae identification
Isolates were identified by amplifying the conserved region of their 16S rRNA genes with universal primers F365 (forward) (5‟-ACWCCTACGGGWGGCWGC-3‟) and R1064 (reverse) (5‟-AYCTCACGRCACGAGCTGAC-3‟) as described by Oezkan et al. (2021) and sequenced using Applied Biosystems. The sequences were then aligned with those available in the National Centre for Biotechnology Information (NCBI) database using the search tool BLAST to determine sequence similarity (http://ncbi.nlm.nih.gov/ BLAST).
Characterization of probiotic properties
Survival under in vitro gastric digestions
Simulated gastric digestion (GD) was performed with minor modifications to the method of Bonatsou et al. (2018). A buffer with a pH of 2.0 (adjusted with 1M HCl) containing gastric solution, NaCl (2.05 g L -1), KH2PO4 (0.60 g L -1), CaCl2 (0.11 g L -1) and KCl (0.37 g L -1) was prepared and autoclaved at 121 °C for 15 min. Pepsin (0.0133 g L -1) and lysozyme (0.01 g L -1) were added before use. Bacterial cultures were grown at 37 °C for 24 h and centrifuged at 7245 x g and 4 °C for 10 min. The pellet was washed without enzymes with the above buffer (pH 2.0). The cells were then resuspended in stomach solution. The OD value was set to 0.5 at 600 nm. Incubation was done on a shaker for 2.5 hours at 37 °C and 200 rpm to simulate peristaltic movements. Samples were counted at the beginning and end of each gastric digestion with MRS agar using the shaker plate method.
Survival under in vitro pancreatic digestions
Simulated pancreatic digestion (PD) was performed with minor modifications to the method of Bonatsou et al. (2018). PD was formulated with bile salts (3.0 g L -1) and pancreatin (0.1 g L -1) in a buffer with a pH of 8.0 (adjusted with 1M HCl) consisting of Na2HPO47H2O (50.81 g L -1) and NaCl (8.5 g L -1). The harvested cells from the previous GD step were washed with Ringer's solution and resuspended in the same volume of simulated pancreatic juice. The OD value was set to 0.5 at 600 nm. Incubation was for 3.5 hours at 37 °C and 200 rpm on a shaker to simulate peristaltic movements. Samples were counted at the beginning and end of each gastric digestion by the pour plate method using MRS agar.
Hydrophobicity
Surface hydrophobicity was measured according to Solieri et al. (2014) with some modifications. Bacterial cultures were grown at 37 °C for 24 hours and centrifuged at 7245 x g for 10 minutes at 4 °C. The pellets were washed twice with PBS and re-suspended to obtain a final OD of 0.5. The absorbance of the suspension was measured at 600 nm. 3 mL of the cell suspension was mixed with 1 mL of xylene (apolar solvent), chloroform (acid solvent) and ethyl acetate (basic solvent) and allowed to stand at 37 °C for 30 minutes. After incubation, the aqueous phase was removed and the absorbance was measured at 600 nm.
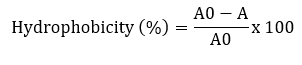
A and A0 are the absorbance values of the aqueous phase after and before solvent addition, respectively.
Auto-aggregation
The auto-aggregation was tested according to the methods described by Somashekaraiah et al. (2019) with minor modifications. Bacterial cultures were grown at 37 °C for 24 hours and centrifuged at 7245 x g at 4 °C for 10 minutes. The pellets were washed twice with PBS and resuspended for a final OD of 0.5. The resulting suspensions were mixed for 15 seconds and incubated at 37 °C. The optical density of the supernatant (OD) was measured after 24 hours at 600 nm.
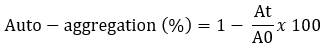
A 0 represents the absorbance of the mixture at t = 0 and At represents the absorbance of the mixture after 24 h of incubation.
Co-aggregation
The coaggregation ability of Lactobacillaceae spp. was tested against the tree pathogens (Escherichia coli ATCC 25922, Listeria monocytogenes ATCC 7644 and Staphylococcus aureus ATCC 25923) according to Jena et al. (2013) with minor modifications. The overnight cultured Lactobacillaceae spp. and tree disease pathogens were centrifuged (7245 x g for 10 minutes at 4 °C) and washed twice. The pellets were resuspended to obtain a final OD600 nm of 0.5. For the coaggregation experiment, equal volumes (2 mL) of the two cell suspensions were mixed and shaken for 10 seconds; absorbance was measured after 24 hours. Control tubes contained a 2 mL suspension of each bacterial strain. The percentage of coaggregation was calculated according to the following formula (Ogunremi et al., 2015), where A represents the absorbance, x and y represent the bacterial cultures and the test bacteria and the (x + y) mixed culture.
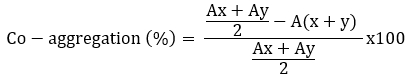
Antimicrobial activity
The strains were inoculated in MRS broth and incubated at 37 °C for 24 hours. The cell-free supernatants were collected (7245×g, 10 min). To assess the potential production of bacteriocin-like inhibitors in the collected supernatants, they were neutralised to pH 6.5 (5 M NaOH). The supernatants were filtered using 0.2 µm sterile Millipore philtres to assess antimicrobial activity. The antimicrobial activities of the pH-neutralised supernatants were evaluated using the agar diffusion assay method of Schillinger and Luecke (1989). E. coli ATCC 25922, L. monocytogenes ATCC 7644 and St. aureus ATCC 25923, pathogenic bacteria, were grown at 37 °C overnight. The suspension of the pathogenic strains (100 µL) was added to the prepared agar, mixed thoroughly and then poured. Supernatants of Lactobacillaceae spp. were added (15 µL) to each well (8 mm diameter) previously cut into agar plates. The plates were incubated at 37 °C for 24 hours. The antimicrobial activities against pathogenic bacteria were recorded as clear zones (mm) around the wells, which was considered as bactericidal activity. If the area around the wells was clearly visible on the agar plate, the result was considered bactericidal activity.
Cholesterol removal
Removal of cholesterol with Lactobacillaceae isolates was performed using a variation of the method described by Tomaro Duchesneau (2014). MRS Broth containing 3 mg/mL bovine bile and 2 mg/mL sodium thioglycolate was prepared for analysis. Water-soluble cholesterol was filtered through a 0.45 µm filter and added to sterile medium (100 µg/mL). 10 mL MRS broth containing 100 µL of cholesterol from each culture grown for 24 hours was added to the broth and incubated at 37 °C for 24 hours. After incubation, cultures were centrifuged at 8000 x g for 10 minutes at 4 °C. Then 1 mL of the resulting supernatant was mixed with 1 mL of potassium hydroxide (KOH, 330 g/L) and 2 mL of ethanol and shaken for 1 minute. The resulting mixture was incubated at 37 °C for 15 minutes and then cooled to room temperature. Then 2 mL distilled water and 3 mL hexane were added to this mixture. The mixture was shaken for 1 minute and incubated at room temperature for 15 minutes. After phase separation, 1 mL of the hexane layer was pipetted into a tube and the solvent evaporated at 60 °C. After drying, 2 mL of o-phthalaldehyde and 500 µL of sulphuric acid were added, mixed homogeneously and incubated at room temperature for 20 minutes. The absorbance was measured at 570 nm. Using different cholesterol concentrations (MRS), a standard curve was constructed (0-500 μg/mL). The percentage of cholesterol removal was calculated according to the following formula:
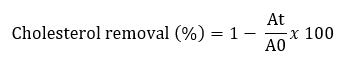
A0 represents the absorbance of the mixture at t = 0, At represents the absorbance of the mixture after 24 h of incubation.
Antibiotic susceptibility
Antibiotic susceptibility of Lactobacillaceae strains was supplemented with various antibiotics including ampicillin, chloramphenicol, erythromycin, tetracycline and gentamycin (Amresco, USA). Bacterial cultures were inoculated overnight at 105 CFU mL -1 in Mueller-Hinton broth. The minimum inhibitory concentration (MIC) for each antibiotic was assessed as the lowest concentration at which no growth was observed after a 24-h incubation period at 37 ºC. The interpretation of the susceptibility status was determined according to the EFSA recommended microbial cut-off values for the assessment of bacterial antibiotic resistance (EFSA, 2012). The EFSA cut-off values for lactobacilli: Ampicillin (10 µg L -1), Chloramphenicol (30 µg L -1), Erythromycin (15 µg L -1), Tetracycline (30 µg L -1) and Gentamicin (10 µg L -1), Kanamycin (30 µg L -1), Streptomycin (10 µg L -1) and Clindamycin (10 µg L -1).
Hemolytic test
The haemolytic activity of the Lactobacillaceae strains was tested on blood agar (containing 7 % (v/v) sheep blood) according to the methods of Leite et al. (2015). All plates were incubated at 37 °C for 48 hours. After incubation, the plates were examined for β-haemolysis, α-haemolysis and non-haemolytic activities.
Antioxidant activity potential
Preparation of intact cells and intracellular cell-free extracts (ICFE)
Antioxidant activities of Lactobacillaceae spp. were measured in two ways: intact cells and ICFE. To measure the antioxidant activities of intact cells, overnight cultured Lactobacillaceae spp. were centrifuged first (5000g, 10 minutes, 4 °C). The resulting pellets were washed twice and re-suspended in PBS buffer (pH 7.4, OD 600nm 0.400±0.05). In order to measure ICFE antioxidant activity, the intact cell suspension was ultrasonically lysed (30 minutes at 37 kHz). Cells were then separated by centrifugation (5000 g, 10 min, 4 °C) and the resulting supernatants were evaluated as ICFE (Cizeikiene and Jagelaviciute 2021).
DPPH radical scavenging activity
DPPH radical scavenging activity of Lactobacillaceae spp. was determined according to Son et al. (2018). Briefly, intact cells or ICFE (2 mL) were mixed with 2 mL of freshly prepared DPPH (0.2 mM in methyl alcohol) and kept in the dark for 30 minutes and centrifuged (5000g, 10 minutes). The absorbance of the resulting supernatant was measured at 517 nm. As a control, water was used instead of the sample. The % DPPH radical scavenging ability was determined as follows:
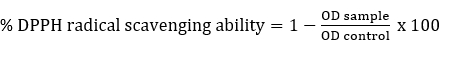
Statistical analysis
Statistical analysis of data was carried out using SPSS (Ver. 19.0 SPSS, Chicago, IL, United States). All the experiments were performed in three independent replicates and data were expressed as means ± standard deviations. The results of parallel studies were carried out by analyzing variance (ANOVA).
Results and discussion
Identification of Lactobacillaceae isolates
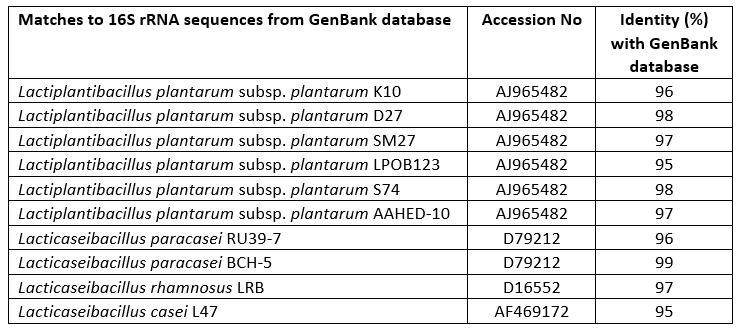
Lactobacillaceae strains were isolated from traditionally produced Turkish Tulum cheese and then identified as genotypes using MALDI-TOF MS biotyper. The identification of these 10 isolates was confirmed by sequencing of the 16S rRNA gene (Table 1). Bacterial strains belonging to the species L. plantarum (6), L. paracasei (2), L. rhamnosus (1) and L. casei (1) were investigated as probable probiotic sources in the present study.
Table 1. Identified matches of Lactobacillaceae spp. isolates obtained by 16S rRNA gene sequencing analysis from the GenBank database
Survival under in vitro gastric and pancreatic digestions
In this study, the growth abilities of 10 strains were determined under gastric and pancreatic conditions in vitro. Table 2 shows that L. paracasei RU39-7 were the most resistant bacteria under gastric conditions (p<0.05). L. plantarum K10, L. plantarum SM27 and L. plantarum S74 had high survival rates under pancreatic conditions (p<0.05).
Overall, the results support previous findings that survival of Lactobacillaceae strains in gastric and pancreatic juices depends on the origin of the bacteria (dos Santos et al., 2020). Furthermore, not all Lactobacillaceae strains are equally resistant to conditions in the stomach and pancreas (Bhushan et al., 2020). The ability of bacteria to survive in gastric and pancreatic conditions is an important feature of probiotics. Probiotics should be resistant to low pH, as viability usually decreases significantly at pH 2.0 and below (Anandharaj and Sivasankari 2014). The present study showed that 10 lactobacilli isolated from traditional Tulum cheese survived in the stomach and pancreas. It was concluded that Lactobacillaceae isolates have different survival rates under in vitro stomach and pancreas conditions.
In this study, some of the selected strains showed a relatively higher survival rate (63 %) compared to the L. plantarum strains reported by Tokatli et al. (2015). It has been reported that foodborne lactobacilli are less resistant to gut conditions than to gastric juice (Tokatli et al., 2015). In this study, L. plantarum UBLP40 showed a survival rate of 40.5-76.8 % under in vitro gastric digestion, which was comparable to L. plantarum ATCC 8014 (73 %; pH 2.0; MRS broth; 3 h) (Xia et al., 2021).
Table 2. Percentage (%) of hydrophobicity, co-aggregation and auto-aggregation obtained for the Lactobacillaceae strains, and percentage of survival rate for Lactobacillus strains at in vitro simulated gastrointestinal conditions
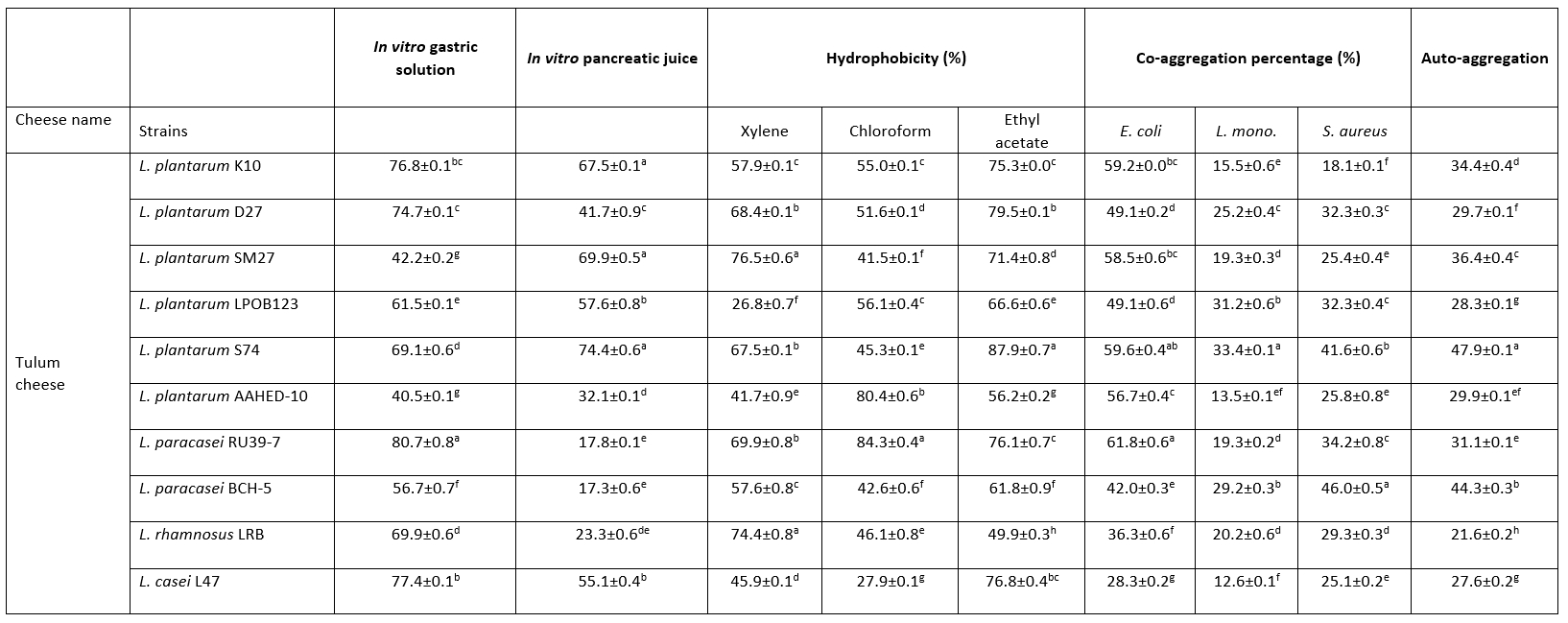
Values with the different superscript lowercase letters in the same column differ significantly (p<0.05)
Hydrophobicity
In this study, the hydrophobicity of Lactobacillaceae strains to xylene, chloroform and ethyl acetate was determined (Table 2). The hydrophobicity of the strains ranged from 26.8-76.5 %, 27.9-84.3 % and 49.9-79.5 % for xylene, chloroform and ethyl acetate, respectively. This study showed the highest cell hydrophobicity rate for strains L. plantarum SM27, L. rhamnosus LRB for xylene, L. plantarum S74 for ethyl acetate and L. paracasei RU39-7 for chloroform (p<0.05). Adhesion of probiotics to the intestinal mucosa is a desirable property. Auto-aggregation and hydrophobicity are prerequisites for the selection of probiotic strains. Thanks to this property, probiotics contribute to adhesion mechanisms and prevent colonisation by pathogens (de Melo Pereira et al., 2018). In this study, 10 Lactobacillaceae isolates showed good abilities to adhere to intestinal cells. The hydrophobicity of the strains ranged from 26.8-76.5 %, 27.9-84.3 % and 49.9-79.5 % for xylene, chloroform and ethyl acetate, respectively. Strains with hydrophobicity above 40 % are considered hydrophobic (Boris et al., 1998). Therefore, our results showed that 10 Lactobacillaceae isolates had high colonisation activity in the gut.
Auto-aggregation
In this study, all Lactobacillaceae strains showed high auto-aggregation abilities. The auto-aggregation values of the Lactobacillaceae strains ranged from 21.6-47.9 % after 24 hours of incubation (Table 2). This study showed the highest auto-aggregation rate for strain L. plantarum S74 (p<0.05).
Auto-aggregation is a method of determining the ability of bacteria to colonise the intestinal wall. The higher the hydrophobicity of the cell surface and the adhesiveness, the easier colonisation of the intestine. Lactobacillaceae strains showed good auto-aggregation and adhesion to xylene, which is considered a measure of cell surface hydrophobicity. Furthermore, adhesion of bacteria to chloroform and ethyl acetate shows electron donor and electron acceptor properties (Bellon-Fontaine et al., 1996).
Co-aggregation
Collado et al. (2008) found that the ability of Lactobacillaceae to co-aggregate is time and strain dependent. In our study, all tested Lactobacillaceae strains showed the ability to co-aggregate with E. coli ATCC 25922, L. monocytogenes ATCC 7644 and S. aureus ATCC 25923. Table 2 shows the results of co-aggregation of Lactobacillaceae strains at 37 °C for 24 h incubation. The results show that the strains L. plantarum S74, L. paracasei RU39-7 against E. coli ATCC 25922, L. plantarum S74 against L. monocytogenes ATCC 7644, L. paracasei BCH -5 against S. aureus ATCC 25923 had the highest co-aggregation ability (p<0.05).
Thanks to the co-aggregation ability, the colonisation of pathogenic bacteria in the human intestine is prevented (Abushelabi et al. 2017). The ability to co-aggregate is an important property of probiotics (Campana et al. 2017). The probiotic properties of LAB increase when the co-aggregation ability of the bacteria increases (Nami et al. 2019).
Table 3. Antimicrobial activities of 10 isolates of Lactobacillaceae
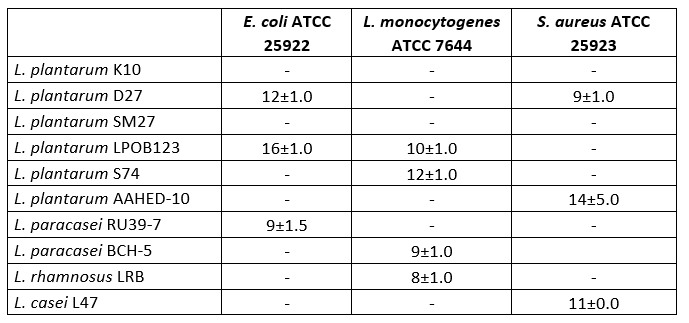
The values show diameters (mm) for inhibition zones
Antimicrobial activity
In the study, tests were carried out on the antimicrobial activity of 10 Lactobacillaceae isolated from cheese. As shown in Table 3, 9 strains were found to have antimicrobial activity. Some Lactobacillaceae strains showed very good antimicrobial activity against E. coli ATCC 25922, L. monocytogenes ATCC 7644 and S. aureus ATCC 25923. The diameter of the inhibition zones ranged from 8 to 16 mm. L. plantarum AAHED -10 showed the strongest antimicrobial activity (14 mm) against S. aureus ATCC 25923 , while L. plantarum LPOB123 also showed strong antimicrobial activity (16 mm) against E. coli ATCC 25922 .
The antimicrobial property of probiotic bacteria is essential for competition with gut pathogens (Jung et al., 2019; Silva et al., 2020). Bacteriocins, organic acids, hydrogen peroxide and surfactants are antimicrobial substances produced by Lactobacillus (Silva et al., 2020). Ten Lactobacillaceae isolates inhibited three types of pathogenic bacteria, including E. coli ATCC 25922 , L. monocytogenes ATCC 7644 and S. aureus ATCC 25923 . Previous reports have demonstrated the antimicrobial activity of Lactobacillus strains against S. aureus and E. coli pathogens (Kumar et al., 2016; Kang et al., 2016). In this study, the L. plantarum D27 strain was found to inhibit the growth of E. coli ATCC 25922 and S. aureus ATCC 25923. Zhu et al. (2015) reported that L. plantarum ZJ217 exhibited bacteriocinogenic activity against S. aureus and E. coli. According to Mulaw et al. (2019), nine Lactobacillus strains showed varying degrees of antimicrobial activity against S. aureus, L. monocytogenes, S. Typhimurium and E. coli. Similar results were found in our study.
Cholesterol removal
In our study, the Lactobacillaceae isolates, especially the isolate L. paracasei BCH-5 (37.9±0.3), were found to have the desired cholesterol-lowering effect (p<0.05). The cholesterol assimilation of the tested bacteria ranged from 17.2±0.2 % to 37.9±0.0 % (Table 4). Tomaro Duchesneau et al. (2014) found cholesterol degradation ranging from 13.13 to 38.99 % in their study. This result is similar to the results we obtained.
The isolates L. plantarum S74 and L. rhamnosus LRB showed the lowest cholesterol degradation rates of 17.4±0.1 % and 17.2±0.2 %, respectively. Even these cholesterol degradation rates were higher than in previous studies.
In the treatment of hypercholesterolaemia, some drugs are used to lower blood cholesterol levels. However, these drugs may have some side effects, such as muscle complications, risk of developing diabetes and gastrointestinal problems (Feingold et al., 2020). Recently, it was found that some strains of lactic acid bacteria can lower low-density lipoprotein cholesterol levels. It was found that several probiotic bacteria of the genus Lactobacillus can be used as cholesterol-lowering agents (Ishimwe et al., 2015). Similar results were found in our study.
Table 4. Cholesterol removal (%) of 10 isolates of Lactobacillaceae
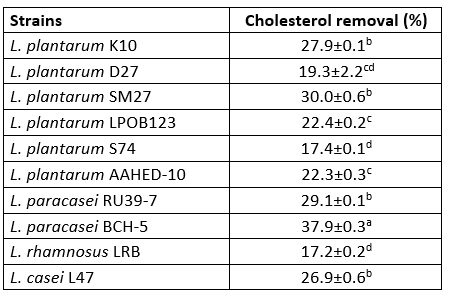
Values with the different superscript lowercase letters in the same column differ significantly (p<0.05)
Hemolytic activity and antibiotic susceptibility
All strains showed no β-haemolytic activity. Five antimicrobials were selected to evaluate the susceptibility of 10 Lactobacillaceae strains. The results of the antibiotic resistance tests showed that all strains isolated from cheese samples were susceptible to most of the antibiotics tested. The antibiotic sensitivity patterns of the Lactobacillaceae strains to different antibiotics are shown in Table 4. The sensitivity patterns of the strains were similar to the different antibiotics. According to the thresholds set by EFSA (2012), all strains tested were found to be sensitive to ampicillin, erythromycin, tetracycline, clindamycin and chloramphenicol. With regard to gentamycin, kanamycin and streptomycin, the Lactobacillaceae were not completely inhibited (Table 5).
Table 5. Results of antibiotic susceptibility for 10 isolates of Lactobacillaceae
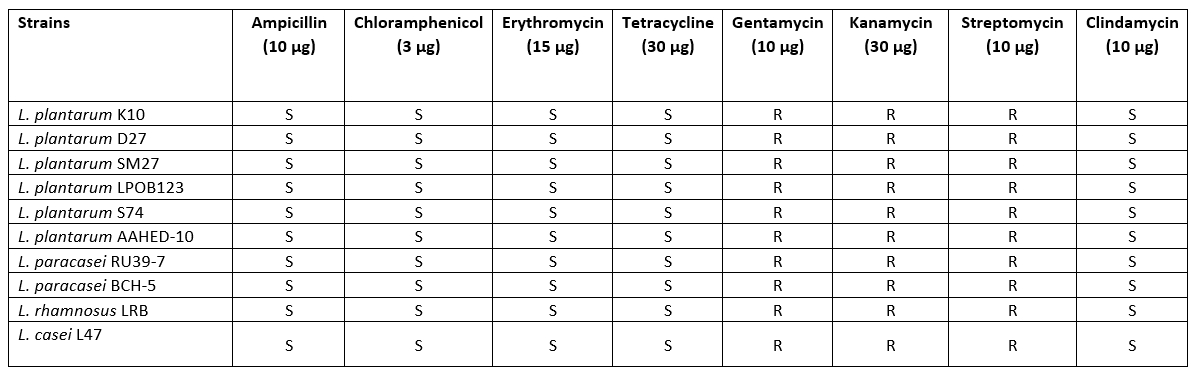
The evaluation of haemolytic activity is one of the criteria for the selection of probiotic strains (Binda et al., 2020). In this study, the Lactobacillaceae strains did not show haemolytic activity, suggesting that the strain is safe to use. The results also support previous studies that Lactobacillus strains are not haemolytic (Bhushan et al., 2020).
One of the most important criteria when selecting probiotics is that they should be safe for human consumption. Probiotic bacteria can become resistant to some antibiotics (de Melo Pereira et al., 2018). Therefore, it is necessary to determine the antibiotic profile of probiotics with regard to clinical risks. All Lactobacillaceae isolates in this study were resistant to gentamicin, kanamycin and streptomycin, but sensitive to tetracycline, ampicillin, erythromycin, clindamycin and chloramphenicol. Similarly, Georgiva et al. (2015) and Amraii et al. (2014) found Lactobacillus strains sensitive to the antibiotics ampicillin, erythromycin and tetracycline. Talib et al. (2019) reported that all Lactobacillus isolates were resistant to vancomycin and gentamicin. In fact, lactobacilli are naturally resistant to vancomycin and gentamicin and do not transfer antibiotic resistance genes between isolates and species (Tokatlı et al., 2015).
Table 6. DPPH radical scavenging activity (%) of 10 isolates of Lactobacillaceae
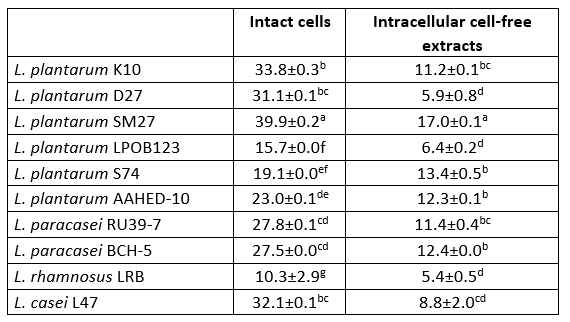
Values with the different superscript lowercase letters in the same column differ significantly (p<0.05)
Antioxidant activity potential
The DPPH scavenging activity of intact cells was higher than that of intracellular cell-free extracts of all bacterial strains tested (Table 6). Intact cells and intracellular cell-free extracts showed radical scavenging activity in the range of 10.3-39.9 % and 5.4-17.0 %, respectively. L. plantarum SM27 intact cells (39.9 ± 0.2 and intracellular cell-free extracts (17.0 ± 0.1) showed the highest DPPH radical scavenging activity (p<0.05). Chen et al. (2014) and Ragul et al. (2020) reported that intact cells had higher DPPH scavenging activity compared to intracellular cell-free extracts.
The antioxidant activity of probiotic bacteria has many benefits. They can prevent gastric ulcers, obesity, diabetes, cardiovascular and chronic diseases, etc. (Dumitrescu et al., 2018). Probiotics can act as antioxidants by scavenging free radicals, reducing iron, chelating metals, increasing antioxidant enzyme levels and regulating the gut microbiota (Feng and Wang 2020). In this study, we determined the antioxidant potential of Lactobacillaceae strains by DPPH radical scavenging.
Conclusion
Two Lactobacillaceae strains isolated from traditional Tulum cheese showed good in vitro survival in simulated gastric and intestinal juice. None of the strains were found to have haemolytic activity. Their antibiotic susceptibility profiles showed them to be safe. In addition, these strains showed good antimicrobial activity against some pathogens. They were also found to have antioxidant activity and to break down cholesterol. In this comprehensive study, we found that few Lactobacillaceae strains have promising probiotic potential, of which L. plantarum SM27, L. plantarum S74 and L. paracasei RU39-7 showed the best probiotic properties (they are the most promising probiotic candidates). In these strains, the percentage of coaggregation and auto-aggregation was above 30 %, and they show hydrophobicity and a survival rate of more than 60 %. Important information was obtained on the functional properties of probiotics in traditional cheeses. It can be concluded that traditionally fermented cheeses can be used for screening and isolation of probiotic strains and starter cultures.
Istraživanje nekih probiotičkih svojstava sojeva Lactobacillaceae izoliranih iz tradicionalnog Tulum sira u Turskoj
Sažetak
Cilj ovog istraživanja bio je ispitati probiotička svojstva sojeva Lactobacillaceae izoliranih iz sira Tulum u Turskoj. Deset sojeva roda Lactobacillaceae spp. su taksonomski identificirani kao Lactiplantibacillus plantarum subsp. plantarum (6), Lacticaseibacillus paracasei (2), Lacticaseibacillus rhamnosus (1) i Lacticaseibacillus casei (1). Ispitana su njihova probiotička svojstva, antimikrobno djelovanje i tolerancija na simulirana gastrointestinalna stanja, uključujući nizak pH, pepsin, pankreatin i žučne soli. Prema dobivenim rezultatima, 10 sojeva bakterija mliječne kiseline pokazalo je visoka autoagregacijska, koagregacijska i hidrofobna svojstva. U svrhu procjene sigurnosti izoliranih sojeva, određen je rezistencija na antibiotike i hemolitičku aktivnost. Također su utvrđeni izolati koji imaju visoku antimikrobnu aktivnost i sposobnost uklanjanja kolesterola, a pokazalo se i da djeluju antioksidativno. Stoga su navedenih izolirani sojevi bakterija mliječne kiseline pokazali obećavajući potencijal za daljnje korištenje kao probiotički sojevi. U ovoj opsežnoj studiji zaključeno je da L. plantarum SM27, L. plantarum S74 i L. paracasei RU39-7 pokazuju najbolja probiotička svojstva. Dobivene su važne informacije o probiotičkim i funkcionalnim svojstvima sojeva Lactobacillaceae izoliranih iz tradicionalnog fermentiranog sira Tulum, koji se mogu koristiti kao bogat izvor probiotičkih bakterija. Također je utvrđeno da tradicionalni fermentirani sirevi mogu služiti kao izvori probiotičkih bakterija.
Ključne riječi: Lactobacillaceae; probiotičke bakterije; tradicionalni sir; Tulum sir
References
dos Santos, L.E., Ginani, V.C., de Alencar, E.R., Pereira, O.G., Rose, E.C.P., do Vale, H.M.M., Pratesi, R., Hecht, M.M., Cavalcanti, M.H., Tavares, C.S.O. (2020): Isolation, identification, and screening of lactic acid bacteria with probiotic potential in silage of different species of forage plants, cocoa beans, and artisanal salami. Probiotics and Antimicrobial Proteins 13, 173-186 . https://doi.org/10.1007/s12602-020-09679-y.
Tomaro-Duchesneau, C., Jones, M.L., Shah, D., Jain, P., Saha, S., Prakash, S. (2014): Cholesterol assimilation by Lactobacillus probiotic bacteria: an in vitro investigation. BioMed research international 1-9.
