INTRODUCTION
The azetidin-2-ones, also well known as β-lactams, are four-membered cyclic amides (1, 2). They are an important class of compounds in medicinal chemistry due to their numerous therapeutic applications. The β-lactam ring is the key structural component of the most widely used class of β-lactam antibiotics (Fig. 1) (3), such as penicillins (4), cephalosporins (5), carbapenems, monobactams and penems (1, 6–8), which have been widely employed chemotherapeutic agents to treat bacterial infections in humans (9) and other microbial diseases (8, 10). In addition to their antibiotic activity, β-lactams demonstrate numerous other interesting pharmacological activities (10–14), such as cholesterol absorption inhibitors (15), thrombin inhibitors (16), human cytomegalovirus protease inhibitors (17), tryptase and chymase inhibitors (12), and vasopressin V1a antagonists (18). β-lactams also exhibit anticancer (19, 20), antitubercular (21), anti-HIV (22), anti-inflammatory (23), antihyperglycemic (24), antimalarial (25), and other biological activities (11, 12).
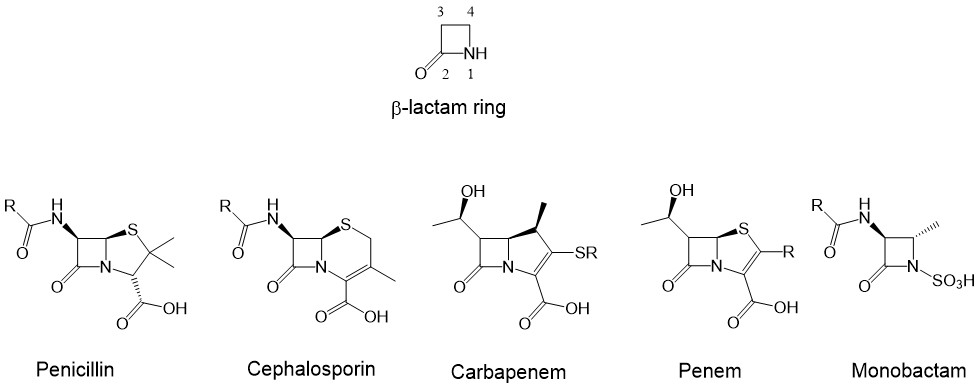
Fig. 1. Structure of β-lactam ring and the representatives of the five major categories of β-lactam antibiotics.
Many methods have been reported in the literature for the synthesis of β-lactams (11, 26, 27). The Staudinger reaction, [2+2] ketene-imine cycloaddition reaction, is the most frequently used method for the synthesis of β-lactam derivatives since the starting materials are readily available (12, 28). This reaction involves cyclocondensation between ketene, generated in situ by treatments of acid chloride (or equivalent) with an imine, in the presence of a mild base (tertiary amine) (28, 29). The reaction produces two new stereocenters (C3 and C4 in the β-lactam ring), so the β-lactam product can be cis-, trans- or a mixture of cis- and trans- isomers depending on the reactants and reaction conditions (11, 28, 30). The Staudinger reaction involves a sequential formation of the N1–C2 and C3–C4 covalent bonds of the β-lactam ring (26, 31).
Another common method of synthesising β-lactams is ester enolate-imine cyclocondensation, also called the Gilman-Speeter reaction (11, 31). However, other notable methods are sometimes employed, including the Kinugasa reaction (26, 32), photoinduced rearrangements (33), oxidative amidation (26, 34), and radical cyclisations (35).
Recently, we reported on the synthesis, separation, and absolute configuration determination of 3-amino-β-lactams (36). In that study, 3-amino-β-lactams were prepared by ester enolate/imine cyclocondensation. This method afforded diastereomerically and enantiomerically pure chiral 3-amino-β-lactams possessing p-fluorophenyl and p-methoxyphenyl substituents at N1 and C4 positions, having a similar structure as ezetimibe, the cholesterol absorption inhibitor.
In the present study, a series of novel (±)- trans-β-lactam ureas 4a‒t were synthesized and evaluated for their antibacterial and antifungal activities in vitro. The compounds 4a, 4b, 4c, 4f, 4h, 4n, 4o, 4p, and 4s were evaluated for their antiproliferative activity in vitro against normal and several types of cancer cell lines. In addition, drug-likeness, ADMET prediction, and biological profiling studies were carried out using in silico tools.
EXPERIMENTAL
General
All needed chemicals were purchased from commercial suppliers Merck (Germany), Fluka (Switzerland) and Kemika (Croatia). Dichloromethane, ethanol, and acetonitrile were dried prior to use according to standard methods. HPLC-grade acetonitrile and methanol were purchased from Honeywell (Germany). Thin layer chromatography (TLC) was carried out on silica gel F254 analytical sheets (DC-Alufolien-Kieselgel F254, Merck KGaA, Germany). Visualization of TLC plates was carried out under ultra-violet irradiation (254 nm) or by ninhydrin. Flash column chromatography was carried out using silica gel Si60 (particle size, 0.04–0.063 mm, 230–400 mesh (Merck).
Melting points were determined using an Olympus BX51 polarising microscope equipped with a Linkam TH600 hot stage and PR600 temperature controller and the Olympus C 5050 ZOOM digital camera (Olympus, Japan).
1H and13C NMR spectra were recorded on Bruker AV 600 (1H 600 MHz and13C 151 MHz) spectrometers (Bruker Technologies, Germany) in deuterated chloroform (CDCl3) at ambient temperature. Chemical shifts (δ) were expressed in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. All of the coupling constants (J) are given in hertz (Hz). Splitting patterns were reported as s, singlet; d, doublet; t, triplet; p, pentet; h, heptet; m, multiplet; bs, broad singlet; dd, doublet of doublets and tdd, triplet of doublet of doublets.
FTIR-ATR spectra were recorded using a Fourier-transform infrared attenuated total reflection PerkinElmer UATR Two (PerkinElmer, USA) spectrometer in the range 450 to 4000 cm–1.
Electrospray ionisation quadrupole-time-of-light high-resolution mass spectrometric (ESI-Q-TOF-HRMS) experiments were performed with Agilent 1290 infinity II/6550 Q-TOF (Agilent Technologies, Germany) instrument.
RP-HPLC analyses were performed on an Agilent 1200 Series System (Agilent Technologies, Germany), equipped with a vacuum degasser, a quaternary pump, a thermostated column compartment, an autosampler, and a variable wavelength detector. RP-HPLC analysis for the determination of the cis/trans ratio of 3-phthalimido-β-lactam 2a/ 2b and analysis of compound 3-amino-β-lactam 3 was performed on Symmetry C18 (150 × 4.6 mm, 5 μm, Waters, USA) column. The method used was as follows: water (eluent A), acetonitrile (eluent B); 0‒15 min gradient: 30 to 70 % eluent B; 15‒18 min isocratic: 70 % eluent B; 18.01‒21 min isocratic: 30 % B; flow: 1 mL min−1, detection wavelength: 254 nm, column temperature: 30 °C, injection volume: 20 μL.
RP-HPLC analyses of racemic trans-β-lactam ureas 4a‒t were performed on a Synergi Polar-RP 80A (150 mm × 4.6 mm, 4 μm, Phenomenex, USA) column using a gradient elution of (A) water + 0.1 % trifluoroacetic acid and (B) acetonitrile as follows: 0‒10 min, 50‒100 % B; 10‒13 min, 100 % B; 13.01‒17 min 50 % B; flow rate 1.0 mL min−1. The column temperature was maintained at 30 °C and the sample injection volume was 20 μL. The UV detection was carried out at 254 nm.
Synthesis of (4-fluorophenyl)[(4-methoxyphenyl)methylene]imine ( 1)
To a solution of 4-methoxybenzaldehyde (6 mL, 49.313 mmol) in dry dichloromethane (30 mL) at room temperature, 4-fluoroaniline (4.67 mL, 49.313 mmol) was added, followed by activated 4-Å molecular sieves powder (2 g). The reaction mixture was stirred at room temperature for 20 h. Then the mixture was filtered through a short layer of Celite (rinsed with dichloromethane) to remove the molecular sieves. The filtrate was concentrated under reduced pressure to afford crude imine as oil. Pure imine 1 (8.1 g, 73 %) was obtained as white crystals after recrystallization from ethyl acetate and hexane.
Synthesis of cis /trans -3-phthalimido-β-lactam ( 2a/ 2b)
A suspension of phthalimidoacetic acid (4.03 g, 19.629 mmol) in dry dichloromethane (150 mL) was cooled to 0 °C under an argon atmosphere. 2-Chloro-1-methylpyridinium iodide (6.69 g, 39.258 mmol) was then added, followed by triethylamine (10.9 mL, 78.516 mmol). The mixture was stirred for 2 hours maintaining the same temperature. Imine 1 (3.0 g, 13.086 mmol) in dry dichloromethane (25 mL) was then added dropwise to this mixture at the same temperature, and stirring was continued for another 2 hours at 0 °C to room temperature. After that, the mixture was refluxed for 20 hours and monitored by TLC and HPLC. After completion of the reaction, the reaction mixture was washed with saturated sodium bicarbonate solution (150 mL), saturated sodium chloride solution (150 mL) and deionized water (150 mL). The organic layer was dried over anhydrous sodium sulfate, filtered, and evaporated to obtain the crude product. The cis/ trans ratio in the crude product was 1:5. Column chromatography over silica gel was performed using chloroform/dichloromethane/ethyl acetate mixture (7:1:1) to isolate cis- and trans-3-phthalimido-β-lactams in pure forms.
Synthesis of 3-amino-β-lactam ( 3)
Ethylenediamine (1.21 mL, 18.156 mmol) was added to a suspension of trans-3-phthalimido-β-lactam ( 2b) (3.78 g, 9.078 mmol) in dry ethanol (100 mL) under argon atmosphere. The mixture was stirred at 65 °C and the progress of the reaction was monitored by TLC. After completion of the reaction (1 hour), the ethanol was evaporated under reduced pressure, and the crude mass was dissolved in ethyl acetate (200 mL). The solution was washed with saturated sodium chloride solution (100 mL) and deionised water (100 mL). The organic layer was dried over anhydrous sodium sulfate, filtered, and concentrated under reduced pressure to obtain the crude product which was then purified by column chromatography over silica gel using ethyl acetate as the eluent solvent. The product 3 was obtained as a white solid.
General procedure for the synthesis of (±)-trans -β-lactam ureas ( 4a ‒ t )
To a solution of (±)- trans-3-amino-β-lactam [(±)- 3] (50 mg, 0.175 mmol) in dry acetonitrile (5 mL) the appropriate isocyanate ( 4a‒ t) (0.263 mmol) was added and the reaction mixture was stirred for 20 hours at room temperature. Then the solvent was concentrated under reduced pressure and the obtained crude product was further purified by column chromatography over silica gel using hexane/ethyl acetate = 4:1 as the eluent. The pure trans-β-lactam ureas were obtained after trituration with hexane in good to excellent yields (75 ‒98 %). The structure of target compounds 4a‒ t was ascertained by spectroscopic methods such as FT-IR,1H NMR,13C NMR, and mass spectrometry.
Physicochemical and spectral data for compounds 1, 2, 3, 4a‒ t are given in Tables I and II. The1H NMR,13C NMR, FT-IR spectra, and HPLC chromatograms of compounds 1, 2, 3, 4a‒ t are shown in the Supplementary materials (Figs. S1 to S112).
Antibacterial activity
The antibacterial activity of the synthesized compounds was evaluated by broth microdilution assay according to the CLSI guidelines (37) with minor modifications using Escherichia coli NCTC 12241 (National Collection of Type Cultures, Salisbury, UK), Staphylococcus aureus ATCC 6538 (American Type Culture Collection, Manassas, Virginia, USA), and Pseudomonas aeruginosa NCTC 12903 as indicator strains. Referent compounds used in the laboratory were: chloramphenicol (Merck&Co., USA), norfloxacin (Merck&Co.), and amikacin (Merck&Co.).
Stock solutions were made at 6.4 mg mL−1 ( m/ V) using DMSO and were diluted with cation-adjusted Mueller Hinton broth (CAMHB) culture media to be tested at a final concentration of 128–0.125 µg mL−1 (prepared by serial two-fold dilutions along the rows of a microtitre plate). Fresh overnight growth of bacteria at 37 °C on tryptic soy agar was used to prepare inocula of 0.5 MacFarland concentration by turbidity adjustment in sterile 5 mL physiological saline (0.85 % NaCl). Inocula were subsequently diluted with CAMHB to reach a final concentration of 5 × 104 CFU per well of a microtitre plate in a total volume of 100 µL per well. All tests were performed on two independent occasions each time in technical duplicates. Each experimental run included positive (drug-free and sample-free wells, i.e., bacteria and sterile media only) and negative (sterile media only) controls and quality control using chloramphenicol. All assays were performed at 35 °C aerobically. Results were interpreted both visually and spectrophotometrically at 600 nm using a microplate reader, and minimal inhibitory concentrations ( MIC) (38), if observed, were screened by culture isolation from the wells, including any available higher concentrations, for evaluating minimal bactericidal concentration ( MBC).
Antifungal activity
Antifungal activity of newly synthesized trans-β-lactam ureas 4a‒ t was determined by the method of microdilution in broth according to CLSI guidelines (39) on strains of Candida albicans ATCC 90028 and Saccharomyces cerevisiae BY 4741 ATCC 4040002. All synthesized trans-β-lactam ureas 4a- t were prepared for antibacterial activity testing except that the YPD broth and agar culture media used were incubated for 48 hours at 30 ± 1 ° C and the wells of the microtiter plate were inoculated at a concentration of 5 × 103 CFU mL−1. The antifungal agent nystatin was used for quality control.
Antiproliferative activity
Antiproliferative activity was studied on selected compounds 4a, 4b, 4c, 4f, 4h, 4n, 4o, 4p, and 4s (Fig. S113). The reason for this selection is due to the fact that only selected β-lactam ureas can undergo further transformation into 5-membered hydantoins, our next synthetic targets currently in preparation. The above-mentioned compounds were tested on three human cell lines: HepG2 (liver hepatocellular carcinoma, HB 8065, from ATCC), A2780 (ovarian carcinoma, 93112519, from ECACC, Porton Down, UK), MCF7 (breast adenocarcinoma, HTB-22, from ATCC) and one human non-tumor cell line HFF1 (untransformed human fibroblasts, SCRC-1041, from ATCC) by the MTT assay (40, 41). The cells were cultured as monolayers and maintained in different media [Dulbecco’s modified Eagle’s medium (DMEM) or Roswell Park Memorial Institute (RPMI-1640) medium] containing different concentrations of fetal bovine serum (FBS). The A2780 cancer cells were cultured in RPMI-1640 medium with 10 % FBS. The MCF7 carcinoma cells were cultured in DMEM supplemented with 10 % FBS and insulin (0.01 mg mL–1). The HepG2 cell lines were cultured in DMEM supplemented with 10 % FBS and HFF1 cells in DMEM containing 15 % FBS. All cell lines were maintained in a humidified atmosphere with 5 % CO2 at 37 °C.
The growth inhibition activity was assessed according to the slightly modified procedure of the National Cancer Institute, Developmental Therapeutics Program (42). The cells were inoculated onto standard 96-well microtiter plates on day 0. The cell concentrations were adjusted according to the cell population doubling time (PDT): 9.6 × 103 per mL for HepG2, A2780, 1.3 × 104 per mL for MCF7, and 1.6 × 103 per mL for HFF1 cell lines. Stock solutions of all the compounds were prepared in DMSO at a concentration of 0.04 mol L–1. The following day, the cells were treated with various concentrations of compounds 4a, 4b, 4c, 4f, 4h, 4n, 4o, 4p, and 4s (from 10–8 to 10–4 mol L–1 in duplicate), and incubated for further 72 hours at 37 °C with 5 % CO2. The working dilutions in DMSO were freshly prepared on the day of testing. After 72 h of incubation, the cell growth rate was evaluated by performing the MTT assay, which detects dehydrogenase activity in viable cells. The MTT cell proliferation assay is a colorimetric assay system, which measures the reduction of tetrazolium component (MTT) into an insoluble formazan product by the mitochondria of viable cells. For this purpose, the substance-treated medium was discarded and 40 µL of MTT reagent was added to each well at a concentration of 0.5 mg mL–1. After four hours of incubation, the precipitates were dissolved in 160 µL of DMSO. The absorbance (A) was measured on a microplate reader at 595 nm (Hidex Chameleon V). The percentage of growth (PG) of the cell lines was calculated according to one or the other of the following two expressions:
if (mean Atest - mean Atzero) ≥ 0, then
PG = 100 × (mean Atest – mean Atzero)/ (mean Actrl – mean Atzero)
or
if (mean Atest – mean Atzero) < 0, then
PG = 100 × (mean Atest – mean Atzero)/ Atzero
where the mean Atzero is the average of the absorbance measurements before the exposure of the cells to the test compound, the mean Atest is the average absorbance after the desired period, and the mean Actrl is the average absorbance after the desired period with no exposure of the cells to the test compound. Each test was performed in quadruplicate in two individual experiments. The results are expressed as IC50, which is the concentration necessary for 50 % inhibition. The IC50 values for each compound were calculated from concentration-response curves using linear regression analysis by fitting the test concentrations that give PG values above and below the reference value ( i.e., 50 %). However, if for a given cell line all of the tested concentrations produce PGs exceeding the respective reference level of effect ( e.g., PG value of 50), then the highest tested concentration is assigned as the default value, which is preceded by a “>” sign.
Drug-likeness and ADMET of compounds 4a‒t
The program ADMET (absorption, distribution, metabolism, excretion, and toxicity) Predictor ver. 9.5 (Simulation Plus Inc., USA) (43) was used to predict the simple structural features: the number of hydrogen bond donors (HBD) and acceptors (HBA), topological polar surface area (TPSA) and physicochemical properties: water solubility (Sw), lipophilicity coefficient (log P), human effective jejunal permeability (Peff) and permeability to Madin-Darby canine kidney (MDCK) cells (Table SII). In addition, the following pharmacokinetic parameters were calculated: percent unbound to blood plasma proteins in humans (hum_fup%), percent unbound in human liver microsomes (fumic%), qualitative estimation of the probability of passing through the blood-brain barrier (BBB), the brain/blood partition coefficient (logBB), the likeness of P-glycoprotein efflux (Pgp sub) and inhibition (Pgp inh) as well as toxicity related parameters (rodent chronic toxicity Rat_and Mouse_TD50 values estimating an oral dose, in mg kg–1 per day) of a compound required to induce tumors in 50 % of a rat/mouse population after exposure over a standard lifetime, qualitative estimation of the likelihood of the hERG potassium channel inhibition in human (hERG_filter), potential toxicity liabilities summarizing all ADMET risk models (Tox_Code). The web tool admetSAR (http://lmmd.ecust.edu.cn/admetSAR2) was used to predict whether a compound may be an inhibitor (CYPs 3A4, 2D6, 2C19, 2C9 and 1A2) or a substrate (CYPs 3A4, 2D6 and 3A4) of the CYP enzymes (Table SII) (44).
Target determination for compounds 4a‒ t by PASS and SwissTargetPrediction
The biological profiles of the new trans-β-lactam ureas 4a‒t were estimated by the machine-learning-based software PASS 2020 (Prediction of activity spectra for substances) (45). The PASS biological screening involved 1,945 PASS-recommended activities including pharmacological effects, mechanisms of action, toxic and adverse effects, antitargets, metabolism-related actions, gene expression regulation, and transporters-related actions. The biological profile is provided as a set of predicted activities along with the associated probabilities of being active (Pa) and inactive (Pi), and with the criteria Pa > Pi and Pa > 0.3, > 0.5 and 0.7 (Table SIII). The SwissTargetPrediction web server (46) was also used to predict potential targets for trans-β-lactam ureas 4a‒ t, based on structural similarity to already known actives. The SMILES (simplified molecular input line entry system) strings of the compounds served as input to all used computational tools.
RESULTS AND DISCUSSION
The synthesized new β-lactam urea structures fall into the category of monobactams, where the β-lactam ring is monocyclic. This moiety is mainly responsible for the antibacterial properties, as it can block bacterial cell wall synthesis through its covalent binding to penicillin-binding proteins (PBPs) (12).
Most chemical modifications commonly made to β-lactam antibiotics involve functionalization of the pharmacophore core, which comprises all four positions of the β-lactam ring. In our study, we have considered the introduction of different substituent groups at positions 2 and 3 (ureido and aromatic) to modulate the possibility of interactions with their targets and consequently achieve new biological activity.
Imine 1 was synthesised according to our recent work; substituents at the N1 and C4 positions of the β-lactam ring are the same as in the ref. 36. Novelty of the current work is restricted to urea moiety and lies in the subsequent derivatization of the NH2 group of β-lactam to yield new structures.
Chemistry
The novel trans-β-lactam ureas 4a‒t were synthesised as indicated in Scheme 1. In the first step, imine 1 was synthesised by a condensation reaction of 4-methoxybenzaldehyde and 4-fluoroaniline in dry dichloromethane in the presence of 4-Å powdered molecular sieves (36). Pure imine 1 was isolated in the 73 % yield after crystallisation from the ethyl acetate/hexane mixture. Imine 1 was then treated with N-phthalimidoglycine in the presence of triethylamine and 2-chloro-1-methylpyridinium iodide (Mukaiyama reagent) to afford a (±)-3-phthalimido-β-lactam ( 2a/ 2b) (47). The crude reaction mixture was analyzed by1H NMR spectroscopy, which clearly showed the presence of two isomeric β-lactams, in a ratio of ca. 1:5 ( cis:trans). Conducted HPLC analysis also corroborates this finding. The1H NMR spectrum of the protons H-3 and H-4 of 3-phthalimido-β-lactam mixture (±)- 2a/ 2b is shown in Fig. S1 (see Supplementary materials).
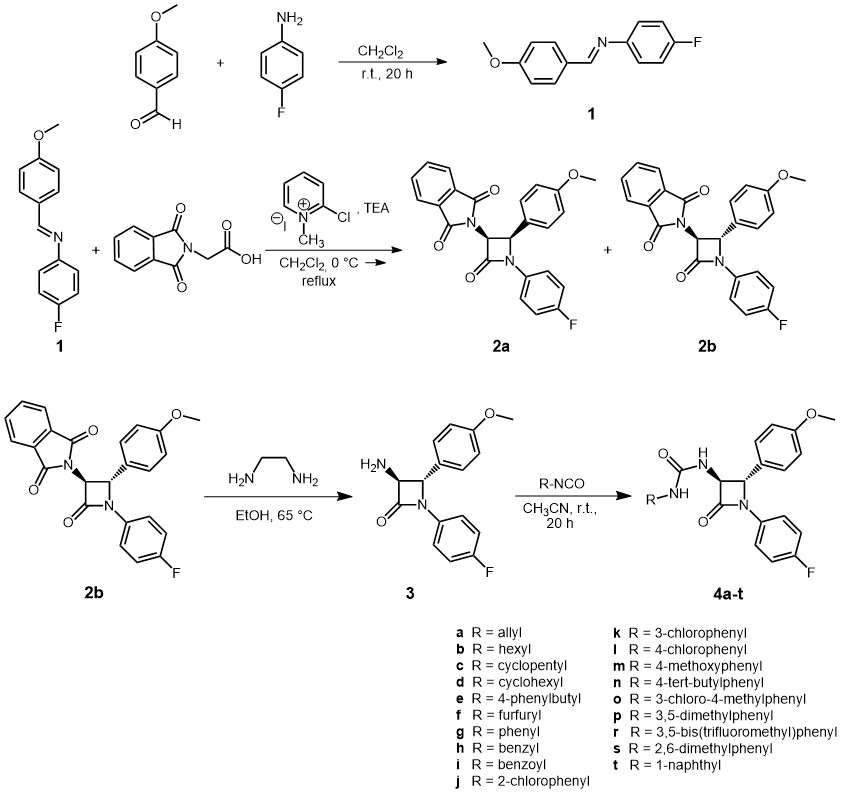
Scheme 1
For the cis stereoisomer, the two protons H-4 and H-3 in the β-lactam ring have appeared as two doublets at 5.42 ppm and 5.65 ppm. The cis vicinal coupling constants were both 5.5 Hz. The corresponding coupling constant for the trans isomer was determined as J = 2.6 Hz, which appeared as two doublets at 5.28 and 5.31 ppm. Based on these data, the structures of the expected diastereomeric β-lactams cis- 2a and trans- 2b were confirmed (Scheme 1). Since the ketene generated in situ from N-phthalimidoglycine and triethylamine is a Sheehan ketene, it has a preference to form trans-β-lactam (28). These diastereoisomers were separated by flash-column chromatography yielding pure trans-β-lactam 2b (75 %) and pure cis-β-lactam 2a (11 %).
In the subsequent step, deprotection of the bulky phthalimide group of compound 2b with ethylenediamine in dry ethanol afforded a free amine, (±)-3-amino-β-lactam ( 3), in good yield (Scheme 1) (47). The treatment of compound 3 with various aliphatic and aromatic isocyanates 4a‒t in dry acetonitrile at room temperature resulted in the isolation of β-lactam ureas 4a‒t (Scheme 1) (48). Products were isolated in good to excellent yields varying from 75 to 98 %. For physicochemical and spectral data see Tables I and II and Figs. S1 to S112.
In the IR spectra of compounds 4a‒ t absorption peaks at 3311–3396 cm–1 (N–H stretch), 2918–2961 cm–1 (aromatic C-H stretch), 2838–2858 cm–1 (aliphatic C–H stretch), 1732–1756 cm–1 (C=O of β-lactam), 1507–1509 cm–1 (ring C=C stretch), 1426 and 1426 cm–1 (C–N stretch), 1246–1255 cm–1 (C–O stretch), 1224–1229 cm–1 (C–F stretch), 1031–1034 cm–1 (saturated C-O stretch) and 830–841 cm–1 (ring vibration due to para-disubstituted benzene) were observed. The IR spectra were dominated by the very strong C=O band, whereas the fingerprint region was characterised by weak IR intensities.
Physicochemical and spectral data of all compounds are given in Tables I and II.
Table I. Chemical names, molecular formula, yields, melting points and R f values of compounds 1, 2, 3, 4a− t
a Eluent: 2a, 2b (chloroform/dichloromethane/ethyl acetate = 7:1:1); 3 (ethyl acetate); 4a − t (chloroform/ethyl acetate = 4:1).
Table II. Spectral data of compounds 1, 2, 3 and 4a–t
a Compound 1 was ascertained by elemental analysis; calcd. C 73.35, H 5.28, N 6.11; found C 73.06, H 5.40, N 6.12.
Antimicrobial activity
All newly synthesised β-lactams derivatives were subsequently tested for their antibacterial and antifungal activities (data not shown). The results showed that compounds 4a‒ t did not inhibit the growth of three indicator bacterial species and two fungal species in the tested range of 128‒0.125 µg mL−1. It is well known that the 2-azetidinone ring is the pharmacophore responsible for the antibacterial and antifungal activity of many structurally diverse compounds (12). It is surprising that the tested compounds did not exhibit antibacterial or antifungal activity. A possible explanation for the experimental results could be found in the substitution at the β-lactam ring. All synthesized β-lactam ureas 4a‒t contain a 4-fluorophenyl group at the N1 position of the β-lactam and a 4-methoxyphenyl group at the C4 position of the β-lactam as well as an alkyl-, aryl-, cycloalkyl- or heterocycle-substituted N-atom of the ureido group attached to the C3 position of the β-lactam ring. The results show that these substituents have no significant effect on the antimicrobial activity.
Antiproliferative activity
The synthesized compounds 4a, 4b, 4c, 4f, 4h, 4n, 4o, 4p, and 4s were tested in vitro for their antiproliferative activity against a panel of three human cancer cell lines HepG2 (liver), A2780 (ovarian), MCF7 (breast), and a healthy cell line HFF1 (human fibroblasts). The results are summarized in Table III. All of the tested compounds show antiproliferative activity against at least two of the three tested cancer cell lines with IC50 values within the range 7.8 ± 8.4 to 82 ± 85 µmol L – 1 . Compound with 3-chloro-4-methylphenyl substituent ( 4o), showed the most potent antiproliferative profile against studied cancer cell lines, with the lowest IC50 value of 7.8 ± 8.4 µmol L – 1 on ovarian carcinoma cell line A2870. However, this β-lactam urea as well as the majority of other tested molecules, is also toxic to normal human skin fibroblasts as well as HFF1 (Table III). The majority of tested compounds were cytotoxic to HFF1 cells. In particular, compounds 4o and 4p exerted the strongest antiproliferative effect on HFF1, while β-lactam ureas 4f, 4n were less toxic to healthy HFF1 cells compared to the cancer cell lines. Compounds 4a and 4s did not show antiproliferative effect on HFF1 nor on HepG2 cells. Taken together, it appears that no selectivity is achieved for compounds 4o and 4p, as they showed toxicity to all cell lines tested. On the other hand, compounds 4a and 4s showed clear antiproliferative activity against the A2780 and MCF7 cell lines, but no activity against the HepG2 and HFF1 cell lines (> 100 μmol L – 1 ). It is important to note that the substitution at the urea moiety proved to be crucial for toxicity and antiproliferative activity, which is not the case for antimicrobial activity. The highest toxicity for all tested cell lines is obtained with the 3,5-dimethylphenyl ( 4p) and 3-chloro-4-methylphenyl ( 4o) substitution at the urea moiety. In contrast, allyl ( 4a) and 2,6-dimethylphenyl ( 4s) substituents have been shown to be the least toxic; surprisingly, only a slight difference in the position of the methyl group in compounds 4p/ 4s showed a major impact on bioactivity.
Table III. In vitro antiproliferative activity of compounds 4a, 4b, 4c, 4f, 4h, 4n, 4o, 4p, and 4s against the HepG2, A2780, MCF7 and HFF1 cell lines determined by MTT assay
Human cancer cell lines: HepG2 (liver), A2780 (ovarian) and MCF7 (breast). Normal cell line: HFF1 (human fibroblast).
aIC50 (concentration that causes 50 % growth inhibition) ± SD (each test was performed in quadruplicate in two individual experiments).
In silico physicochemical and biological profiling
Herein, ADMET properties for the synthesised compounds 4a‒ t were estimated by the program ADMET Predictor 9.5 (43) and the web server admetSAR (44).
All synthesised β-lactam ureas, with the exception of 4r, conform to Lipinski’s and Veber’s rules for drug-like, low molecular mass compounds (Table SI). An orally administered molecule as per Lipinski’s rule of five should have log P ≤ 5, Mr < 500 Da, HBD ≤ 5, and HBA ≤ 10 (49‒51), while according to Veber’s rule (52) its TPSA and the number of rotatable bonds should be less than 140 Å2 (or 12 or fewer HBD and HBA) and 10, resp. The outlier 4r with 3,5-bis(trifluoromethyl) substituent violates Lipinski's rule in terms of molecular mass ( Mr =541.43) and lipophilicity (log P = 5.46). As it is well known, lipophilicity being a very important property of a drug strongly affects its pharmacokinetics, pharmacodynamics, and toxicological profile, i.e., solubility, absorption, distribution, metabolism, excretion, binding to plasma proteins, interaction with the receptor, transport across cell membranes and BBB (53). All β-lactam ureas 4a‒ t are estimated to be non-ionisable compounds with log P lying within the range 2.5–5.0 (except 4r). They are predicted to have moderate water solubility which agrees with our observation (Table SI), and moderate to high membrane permeability (Table SII), which altogether indicates their good oral bioavailability.
Good membrane permeability of a compound may be affected by its interaction with efflux pumps such as P-glycoprotein (Pgp). According to ADMET predictor models, all synthesised trans-β-lactam ureas 4a‒ t are estimated to be Pgp substrates, and many of them are predicted to be Pgp inhibitors (Table SII). The BBB is a selectively permeable membrane that controls the passage of molecules into and out of the brain (54). Considering that β-lactam ureas 4a‒ t are predicted to be Pgp substrates, not penetrate BBB, and have low retention in the brain (BBB and logBB parameters in Table SII, resp.), it is likely that they do not enter the brain. As for the distribution in the body, the synthesised molecules are likely to be transported through the body by plasma proteins (low hum_fup% values in Table SII).
With regard to metabolism, the web server admetSAR was used to estimate the likelihood that the compounds are substrates and inhibitors of cytochrome P450 (CYP450) enzymes. These oxidoreductases belong to a superfamily of heme-containing proteins involved in the phase I metabolism of drugs and other xenobiotics (55, 56). Among CYP450 enzymes, the CYP1, CYP2, and CYP3 families mediate 70–80 % of all phase I metabolic reactions of clinically relevant drugs with CYP3A4 performing at least half of the drug metabolism (57, 58). According to admetSAR predictions, all trans-β-lactam ureas 4a‒ t should be substrates for CYP3A4 (and not for other considered CYPs), and half of them are also predicted to be its inhibitors (Table SII). This also implies the importance of determining cellular metabolites when interpreting in vitro results. The CYP450-mediated intrinsic clearance of β-lactam ureas is also supported by quite low estimated fractions of unbound compounds in human liver microsomes (%Fumic). These compounds are classified as class 2 of the extended clearance classification system (ECCS), with hepatic metabolism being the major route of excretion. Class 2 molecules (high permeability bases/neutral substances) (59) tend to be predominantly metabolized in the liver by enzymes such as CYP3A4. They cross the basolateral membrane of hepatocytes by passive diffusion and are metabolised and excreted in the urine/bile as phase I or II metabolites (60).
The synthetized compounds should have low hepatotoxicity and mutagenicity. The β-lactam ureas 4r and 4t were predicted as the most potentially toxic molecules with potentially having negative cardiotoxic and carcinogenic impacts (Table SII).
The PASS software (45) and SwissTargetPrediction online server (46) were used to envision the biological activities of nineteen trans-β-lactam ureas 4a‒t. By using built-in machine-learning models, PASS simultaneously predicts more than a thousand biological and toxicological activities from the 2D structural formulae of the compounds. On the other hand, SwissTargetPrediction estimates likely protein targets based on structural similarity with 327,719 already known actives on 2,092 human proteins. Although PASS predicted 177 possible activities for at least one of the new β-lactams including treatment of atherosclerosis, and anti-inflammatory and analgesic activity which are consistent with the already known activities of this chemical class , they were predicted with Pa ≤ 0.5 (Table SIII) . Similarly, according to SwissTargetPrediction a list of human proteins, mainly kinases, were suggested as tentative targets with low probability. No predictions were found by either algorithm with high probability as there were no similar chemicals in the training models of PASS and among the SwissTargetPrediction active compounds, indicating a substantial novelty in the structures of the synthesized β-lactam ureas .
Limitations of the study
In this research, compounds involved in synthesis and evaluation aimed to yield new products that possess interesting biological activities. All these compounds have a common chemical β-lactam moiety, which usually has a broad spectrum of activities, especially the antimicrobial activity, which is completely absent in our study. In contrast to this finding, their biological profile included antiproliferative activities. More detailed prospects for future studies on this class of bioactive compounds are expected and will also be extended after the chemical conversion of β-lactam ureas into hydantoin derivatives, possibly as a continuation paper.
CONCLUSIONS
In conclusion, a series of nineteen novel drug-like racemic β-lactam ureas 4a‒ t were synthesised in good to excellent yields. Their biological potential was evaluated in vitro by testing the antimicrobial and antiproliferative activities and in silico by using PASS, SwissTargetPrediction, and ADMET predictor approaches. Neither of the β-lactam ureas racemates showed antibacterial activity against E. coli, P. aeruginosa, and S. aureus and antifungal activity against C. albicans and S. cerevisiae. Most of the tested compounds 4a, 4b, 4c, 4f, 4h, 4n, 4o, 4p, and 4s showed antiproliferative activity on tested tumor cell lines HepG2, A2780, and MCF-7 in the micromolar range, but were also cytotoxic for normal fibroblasts HFF-1. The β-lactam urea 4o with 3-chloro-4-methylphenyl substituent ( IC50 = 7.8 ± 8.4 µmol L–1) was found to have promising antitumor activity against ovarian carcinoma cell A2780; however, compounds 4o and 4p showed a strong cytotoxic effect against normal cell line HFF ( IC50 = 8.0 ± 5.1 µmol L–1 and IC50 = 4.5 ± 8.9 µmol L–1, resp.). In addition, compounds 4a, 4f, 4n, and 4s exerted no or low toxic effects on human fibroblasts HFF-1 in the studied concentration range. In silico ADMET profiling described compounds 4a‒ t as moderately water-soluble and membrane permeable, quite lipophilic molecules with a propensity to be metabolised by CYP3A4 (and excreted hepatically) and expelled by Pgp pump. PASS and SwissTargetPrediction predicted some significant biological activities and targets for compounds 4a‒ t but with low probability, consistent with the novelty of their structure. Considering predicted activities, e.g., atherosclerosis treatment, and low toxicity risk on human fibroblasts, molecules 4a and 4f can be suggested as the most promising for further investigations comprising chemical structure modifications.
Supplementary materials are available upon request.
Acknowledgments. ‒ We would like to thank the NMR Centre for recording the spectra.
Funding. ‒ This study was supported by the Croatian Government and the European Union through the European Regional Development Fund – the Competitiveness and Cohesion Operational Programme (KK.01.1.1.01) through the project Bioprospecting of the Adriatic Sea (KK.01.1.1.01.0002) granted to The Scientific Centre of Excellence for Marine Bioprospecting – BioProCro.
Conflicts of interest . ‒ The authors declare no conflict of interest.
Authors contributions. – Conceptualization, M.J., T.D. and M.R.; methodology, M.J., V.S., K.B. and D.V.; analysis, M.J., K.B. and D.V.; investigation, M.J., V.S., K.B. and D.V.; writing, original draft preparation, M.J., V.S. and K.B.; writing, review, and editing, D.K., T.D. and M.R. All authors have read and agreed to the published version of the manuscript.
