INTRODUCTION
The biological immune system has proved to be a rich source of inspiration for computing[1-15]. Artificial immune systems take inspiration from the immune system to provide powerful metaphors for robust and distributed computing. In this article, I employ an immune system inspired approach to solve a problem in program verification: that of finding a program invariant.
An invariant of a program is a mathematical formula that captures the semantics of the program[16] and is used in automatic program verification. The shape of an invariant is its approximate polynomial representation. Once the shape of the invariant is predicted, deterministic techniques can be used to generate the exact form of the invariant[17]. Hence, the prediction of invariant shape is of paramount importance for program verification.
An artificial immune system algorithmic framework is proposed to carry out the machine- learning task of predicting invariant shape from an instance of a program. Program invariants encapsulate the computability of a particular program, e.g. whether it performs a particular function correctly and whether it terminates or not. We hope this work will also lay the foundation for applying concepts of theoretical incomputability and undecidability to biological systems like the immune system that perform robust computation to eliminate pathogens[8-15].
IMMUNOLOGICAL PRELIMINARIES
A chemical species that can be recognized by the adaptive immune system is known as an antigen (Ag). When an organism is exposed to an Ag, some specialized immune system cells called B cells respond by producing chemicals called antibodies (Ab’s). Ab’s are molecules attached primarily to the surface of B cells whose aim is to recognize and bind to Ag’s. By binding to these Ab’s the Ag stimulates the B cell to proliferate and mature into plasma cells that secrete Ab. An organism is expected to encounter a given Ag repeatedly during its lifetime. The effectiveness of the immune response to secondary encounters is enhanced by the presence of memory cells associated with the first infection, capable of producing high- affinity Ab’s after repeat encounters. Such a strategy ensures that the speed and accuracy of the immune response becomes successively higher after each infection. This gives rise to associative memory where the stored pattern is recovered through the presentation of an incomplete version of the pattern. The repertoire of activated B cells is diversified[18-21] and B-cells with higher affinity for the antigen are selected to enter the pool of memory cells.
AUTOMATED PROGRAM VERIFICATION AND PROGRAM INVARIANTS
The field of automated program verification started with seminal work by Floyd[22] and Hoare[23]. They introduced the concept of a loop invariant: a mathematical formula that remains true throughout the execution of a loop. The loop invariant completely captures the semantics of the loop, and along with the program preconditions and postconditions, can be used to show correctness of the program[23].
Previous work[16] has shown how the loop invariant for a particular program can be generated by a priori agreement on the shape of the invariant: the approximate polynomial representation of the invariant. However, the shape of the loop invariant can be hard to deduce for many programs.
The following shows an example program:
{A 0, B 0}
x : = A;
y : = B;
z : = 0;
while x > 0 do
if odd(x) then z : = z + y;
y : = 2 * y;
x : = x/2;
end while
Assuming the shape of the program invariant as Ishape: Ax + By + Cz + Dxy + Eyz + Fxz + Gxyz + H = 0, (where A, B, C, D, E, F, G and H are constants or program variables), using quantifier elimination[16] the final loop invariant is Ifinal: z + xy - AB = 0. Coupled with a precondition P: {A 0 ^ B 0 ^ x = A ^ y = B ^ z = 0} and a postcondition Q: {z = A*B}, it can be shown that this invariant is consistent with Q i.e. the program correctly multiplies 2 numbers A and B and stores the result in z.
Finding the precise shape of the loop invariant is generally a non-trivial process and the algorithm proposed aims to use “cues” from the program to make informed predictions about the invariant shape and ultimately help in automated program verification.
PROPOSED COMPUTATIONAL FRAMEWORK
Here we propose a computational framework for predicting program invariants. An artificial immune system (AIS) algorithm will be used to generate shapes of program invariant. Initially the AIS will be trained on programs, for which the shape of invariant is known. Then a program will be presented to the AIS and it will try to predict the form of the invariant.
An AIS approach presents many advantages over a traditional Machine Learning (ML) approach. In an AIS, recognition can be sloppy[24] i.e. if it has previously recognized program P (with an invariant I), then a new program P’ “similar” to P, can also be recognized, and an invariant I’ can be generated (that is similar in form to I). This is akin to our immune system recognizing a previously encountered pathogen (program), and generating antibodies (invariant) similar to the previously produced antibodies.
The natural immune system produces antibodies by a process of mutation, and the same process is emulated in AIS algorithms. A candidate solution (invariant) will be generated, and then the solution will be improved by in-silico mutation.
Previously encountered programs and their corresponding invariants will be stored as memory B cells. When a program similar to a stored one is presented, the time taken to generate the invariant will be shorter than the time taken to generate the original invariant (secondary response).
COMPONENTS OF THE ARTIFICIAL IMMUNE SYSTEM
Here we define the specific components of the AIS have to be determined. What is the program analogue of an antigen and an antibody?
A program fragment is defined to be either an assignment statement, a statement containing an iteration construct (for, while, repeat, etc.), or a statement having a conditional check (if <condition> then) e.g. x: = x + 2, and while (x > 0) do, and if (x > 3) then, are all program fragments.
The analogue of an antigen is a program fragment and the corresponding analogue of an antibody is an invariant for the program fragment it recognizes. Hence, the AIS will be presented with an antigen (program fragment), and the immune system cells will either produce the antibody (invariant) immediately if it has encountered this antigen before, or will undergo mutations to generate the correct antibody (invariant).
The individual invariants for each program fragment will then be recombined to generate the invariant for the whole program.
A SHAPE SPACE AND ANTIGENIC DISTANCE FOR PROGRAMS
We need a measure of distance between disparate program fragments, so that the AIS can recognize them and generate an antibody in response. For a natural immune system, the antibody combining region relevant to antigen binding can be specified by a number of “shape” parameters[25] which denote the size and shape of the combining site or physical characteristics of the amino acids.
If there are N shape parameters, they can be combined into a vector, and antibody combining sites and antigenic determinants can be described as points Ab and Ag, in an N - dimensional Euclidean vector-space called shape space[25].
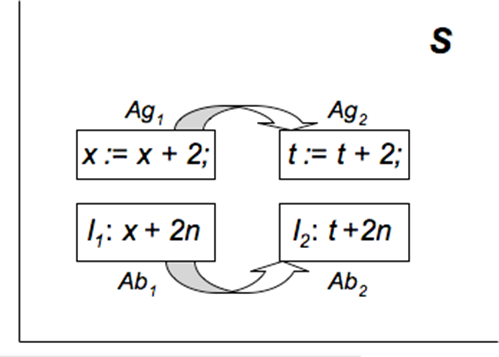
Antigenic distance between 2 antigens is the distance in shape space[26] between them e.g. ||Ag1 - Ag2|| is the distance between antigens Ag1 and Ag2 in shape space S. The antibody distance is the distance ||Ab1 - Ab2|| in shape space between 2 antibodies Ab1 and Ab2.
I define the program fragment shape space as the N-dimensional Euclidean vector space of program fragment characteristics like identifier name, exponent on the identifier, operator, etc. I define the corresponding program fragment antigenic distance as the distance ||P1 - P2|| between 2 program fragments P1 and P2 in program fragment shape space. The program fragment antibody (invariant) distance is the distance ||I1 - I2|| between 2 program fragments I1 and I2 in program fragment shape space.
Let us consider 2 program fragments P1: x: = x + 2 and P2: t: = t + 2. The corresponding antibody (invariant) for P1 is I1: x = x + 2n, where n is a program variable or constant (since upon n - 1 iterations, x gets the value x + 2n). Let P1 and I1 constitute the training set. Then the AIS should be able to produce an antibody (invariant) for the program fragment P2 even though it has never encountered this antigen (program) before. The correct invariant is I2: t = t + 2n (where n is a program variable or constant) and this is indeed what the AIS generates by somatic hypermutation. The program P1 differs from P2 by 1 mutation (replacing x by t on both sides of the assignment) i.e. the program fragment antigenic distance ||P1 - P2|| is 1. The invariants I1 and I2 also differ by 1 mutation (replacing x by t) i.e. the program fragment antibody (invariant) distance ||I1 - I2|| is 1. Hence, when an AIS trained on (P1, I1) is presented with P2, it produces I2 using one mutation from I1 (Fig. 1).
PROPOSED ALGORITHM
In this section we outline the proposed immune system inspired algorithm. The artificial immune system (AIS) would be trained on the antigen (program fragment) P1: x: = x + 2 and given the antibody (invariant) I1: x = x + 2n as a solution (training phase). The AIS stores the solution I1 as a memory detector.
When an entire program (as opposed to a program fragment) is presented to the AIS, it breaks the program up into program fragments (all the assignment statements in the program), and then “presents” each of these antigens (fragments) to itself.
If an antigen (program fragment) P2 “similar” to P1 is detected, it will generate I1 as a candidate solution. If I1 itself does not act as an invariant, the AIS will keep on carrying out randomly on I1 until it evolves the final antibody (invariant) I2 that will act as the invariant for the program presented (somatic hypermutation phase). This is akin to how the natural immune system mutates B cell receptors and ultimately produces a receptor that can recognize the antigen. The algorithm may also use some heuristics to guide the mutation process e.g. if an antigen (program fragment) of the form p: = p + 5 is encountered, it would search its repertoire for a program fragment that is closest in program shape space to this e.g. x: = x + 5 is closer to the presented antigen (1 mutation) than y: = y + 7 (2 mutations). Additionally, we will have to ensure that each mutation is sound i.e. there is no such mutation that would generate a wrong invariant for the corresponding mutated program fragment. In the last step, the AIS incorporates Ii into its memory pool (learning phase).
The AIS then presents the next program fragment P3, generates the invariant I3 and stores it in the memory population, and so on until all program fragments have been presented. Finally, the AIS combines all invariants linearly, producing a polynomial (shape of invariant) that captures the semantics of the entire program.
RESULTS
The AIS (trained on P1, I1) presented with suites of entire programs would successfully generate the shape of the invariant. The first program is shown below:
(x,y,u,v) : = (a,b,b,0);
x : = a; y : = b;
u : = b; v : = 0;
while (x y) do
while (x > y) do x : = x - y; v : = v + u; end while;
while (x < y) do y : = y - x; u : = u + v; end while;
end while
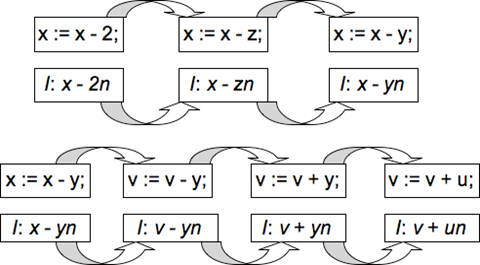
This program takes 2 positive integers a and b, and calculates their g.c.d and l.c.m. The AIS presents itself with each assignment statement sequentially. The first 4 assignment statements (lines 1-2) have no invariant, since they are not contained inside any loop. Hence, the AIS does not generate any invariant for them. The progress of the algorithm on the next 2 assignment statements (x: = x - y; v : = v + u;) is shown below in Fig. 2.
The AIS starts from the training set (P1: x: = x + 2 & I1: x = x + 2n) and then mutates the operators and operands to create the invariant I3: x = x - yn for the program fragment P3: x: = x - y. The AIS stores I3 in the memory population and for the next assignment statement (v: = v + u;), it starts mutating from (P3, I3) until it creates the invariant I4: v = v + un for the program fragment P4: v: = v + u.
For the next set of assignment statements (y : = y - x; u : = u + v;), the AIS then generates the invariants I5: y = y - xn and I6: u = u + vn (not shown). The 4 invariants I1, I2, I3 & I4 are then combined linearly (with n being substituted for all program variables, namely x, y, u, v) to yield the invariant shape Ishape: Ax + Bv + Cy + Du + Exy + Fy2 + Guy + Hvy + Jxu + Ku2 + Lvu + Mx2 + Nvx + Pv2 + Q = 0, where A, B, C, D, E, F, G, H, J, K, L, M, N, P and Q are constants or program variables. This is the correct invariant shape, since using quantifier elimination[16], the final invariant yielded is Ifinal: xu + yv - ab = 0 (with A = B = C = D = E = F = G = K = L = M = N = P = 0, Q = -ab, H = J = 1).
Finally we test the AIS on another standard program[16] shown below:
{A ≥ 0, B ≥ 0}
x : = A;
y : = B;
z : = 1;
while y > 0 do
if odd(y) then y : = y - 1; z : = x * z;
else x : = x * x; y : = y/2;
end while
This program calculates AB and stores it in z. The AIS would calculate the invariant for the program fragment P5: z : = x * z as I5: z = xn * z. For the program fragment P6: x : = x * x, it generates the invariant I6: x = exp(x, exp(2,n)), where exp() is the exponentiation function. Combining all the program fragment invariants, gives us the following invariant shape:
Ishape: Azxx + Bzxy + Czxz + D.exp(x,exp(2,x)) + E.exp(x,exp(2,y)) + F.exp(x,exp(2,z)) +G = 0.
This is the exact shape of the invariants, since quantifier elimination yields the final invariant
Ifinal: zxy = AB (with A = C = D = E = F = 0, G = -AB).
We can now readily verify the working of the program. When the loop terminates, the invariant is true and y = 0, which yields the correct postcondition: z = AB.
The proposed algorithm would use a sequence of mutations, guided by heuristics, to generate the correct invariant for a program invariant.
CONCLUSION AND FUTURE WORK
We have proposed a computational framework for an immune system inspired approach for automated program verification. The immune system inspired algorithm breaks up a program into fragments and presents them to itself. It then generates an invariant in response to each program fragment and ultimately combines them to create the general shape of the invariant. We show how this approach can be used to generate the general form of the program invariant for non-trivial benchmark programs[16].
Future work will focus on theoretical research into whether there are classes of programs
for which a linear combination of individual program fragment invariants might not generate the invariant for the entire program. Another avenue of future investigation would be to look into how mutations on exponentiation would affect the invariant e.g. x : = x + 2 getting mutated to x : = x2 + 2. Lastly, our approach does not consider program fragments having iteration constructs like while, repeat, etc. and future research will investigate how incorporation of such program fragments can enhance the predictive power of the algorithm.
A lot of work has been done on incomputability, undecidability and program termination in theoretical computer science. The best characterization of this comes in the form of the Halting Problem formulated by Alan Turing. Biological systems also perform computing, e.g. the immune system computes the most efficient way to eliminate pathogens in a timely manner without harming the host[8-15]. However it has been more difficult to define incomputability and undecidability for biological systems.
Program invariants encapsulate the computability and correctness of a particular program, e.g. what it does and whether it terminates or not. This work lays the foundation of applying computability to biological systems especially the immune system that performs computation.
The present work also applies immune system inspired algorithms to find program invariants and prove correctness and termination. It is intriguing to speculate that it is also possible to go in the reverse direction and translate the complexities of the immune system into an equivalent computer program. The translated computer program can then be analyzed for mathematical properties of what it computes[27,28]. Hence this work can be extended to provide a theoretical framework for understanding the limits of computation in the immune system.
The present computational framework can be used to account for cases when the immune system fails to clear infections as is the case in certain virulent infections[29].
This approach can also be similarly extended to analyse substrates for computing that are non-silicon based and can be used to probe the computational nature of life itself[15].
In summary, the present work applies the theoretical concepts of undecidability to immuno- computing and possibly biological computing in general. We view this work as the first step towards elucidating the fundamental limits of computing in immunology and possibly biology as well.