Introduction
Programmed cell death (PCD) is generally described as a genetically controlled mechanism that eliminates specific cells under developmental and environmental stimuli. In recent years, due to the complexity of classification based on morphological and biochemical characteristics, plant PCD has been divided into two main categories: developmentally regulated PCD (dPCD) and environmentally induced PCD (ePCD), depending on the type of activation. dPCD is activated by internal stimuli, whereas ePCD is triggered by various biotic and abiotic stress factors such as pathogens, fungi, salinity, UV radiation, drought, heat, flooding, heavy metals, etc. (Li et al. 2007, Yanık et al. 2017, Minina et al. 2021, Paes de Melo et al. 2022).
Probably the most thoroughly understood form of PCD in animals is apoptosis, which is associated with caspase activity and initiated and regulated by these proteases. Although plants lack caspases, PCD is regulated by various proteases such as metacaspases, vacuolar processing enzymes (VPEs), and phytaspases which are suggested as the main regulators of PCD in plants. Moreover, VPEs are also proposed as the primary regulators of vacuolar cell death in plants. Vacuolar cell death is a crucial process in plant development accompanied by striking expansion of lytic vacuole digesting cytoplasmic content, which leads to the self-clearance of dying cells (Emanuele et al. 2018). In many plants, PCD generally shows characteristic features such as cell and cytoplasm shrinkage, chromatin condensation, DNA laddering, decreased mitochondrial membrane potential, disruption of microtubule organization, and cytochrome c (cyt-c) release from mitochondria to the cytoplasm (Huang et al. 2014, Paes de Melo et al. 2022).
In recent years, the increasing presence of consumer products containing nanoparticles (NPs) has raised concerns about nano-waste as a potential environmental threat. Silver nanoparticles (AgNPs) are among the NPs most used in consumer products, posing a potential risk as pollutants. AgNPs are extensively used in many fields due to their antimicrobial features, e.g., in the healthcare industry, food packaging, water treatment, textile coating, cosmetics, etc. (Alshameri and Owais 2022). With the development of nanotechnology, they have also attracted attention as nano-pesticides for controlling bacterial and fungal diseases in agriculture (Chhipa and Joshi 2016). The increasing prevalence of nanotechnology in consumer products inevitably leads to the release of AgNPs into the ecosystem during synthesis, consumption, and disposal processes. This has prompted researchers to assess the eco-toxicological impact of AgNPs on organisms such as human cells, algae, and plants (McGillicuddy et al. 2017). In plants, exposure to AgNPs can induce oxidative stress, genotoxicity, up-regulation of stress-related genes, and cell death, depending on the size and concentration of AgNPs and the plant species (Mirzajani et al. 2014, Huang et al. 2022). Investigations based on NP size have shown that 10 nm-sized AgNPs induce various effects in different plant species. In Triticum aestivum, they inhibit root and shoot length, induce metallothionein gene expression, and cause oxidative stress (Dimkpa et al., 2013). In Arabidopsis thaliana, 10 nm-sized AgNPs disrupt chloroplastic membrane structure, reduce chlorophyll content, and alter the expression of antioxidant and aquaporin-related genes (Qian et al., 2013). Additionally, Wolffia globosa exposed to these AgNPs show higher malondialdehyde (MDA) content indicative of oxidative stress, along with an increase in superoxide dismutase (SOD) activity (Dimkpa et al. 2013). Our previous study also revealed the phytotoxicity of 10 nm-sized AgNPs, with observed adverse effects on root and shoot length, increased H2O2 and MDA content, antioxidant activity, lignin accumulation, and callose deposition (Yanık and Vardar 2019). Moreover, a recent study has demonstrated that AgNP treatment induces autophagy-type PCD in grapevine suspension cells, characterized by reactive oxygen species (ROS) generation, caspase-3-like activity, an increase in poly-ubiquitin conjugated proteins, and degradation of the proteasome 20S subunit PBA1 (Filippi et al. 2019). Despite their toxic behavior, the potential of AgNPs to induce cell death in plant cells remains unclear. The present study aims to evaluate PCD hallmarks induced by 10 nm-sized AgNPs in wheat roots. We investigated various cell death features, including mitotic division, chromosomal behaviors, nucleus morphology, DNA fragmentation, cyt-c release, and caspase-1-like activity, along with the expression of cysteine proteases genes (TaVPE4, TaMCA1, and TaMCA4). Our goal is to understand the potential impacts of AgNPs on agricultural processes.
Plant material and light microscopic observations
Wheat (T. aestivum L. cv Demir 2000) seeds were obtained from the Central Research Institute for Field Crops (Ankara, Türkiye). The seeds were germinated with distilled water and then exposed to different aqueous concentrations (0, 0.5, 1, 5, 10, and 20 mg L-1) of 10 nm-sized AgNPs (Sigma-Aldrich 730785) for 15 days, following the method described by Yanık and Vardar (2019). For screening anatomical alterations, roots were fixed in 6% glutaraldehyde (v/v) in 0.1 M phosphate-saline buffer (PBS, pH 7.8) for 16 h. Semi-thin cross-sections (1 μm) were stained with toluidine blue. To determine the mitotic index (MI) and chromosomal abnormalities (CA), roots were fixed for 24 h with acetic acid: alcohol (1:3) and hydrolyzed with 1 M HCl for 10 min at 60 °C. The roots were then kept in the dark for 30 min with Schiff’s reagent and squashed with 2% aceto-orcein (w/v). Approximately, 1500 cells were scored from each group, and the MI and CA were expressed as the number of the cells in mitosis per 100 cells and expressed as a percentage.
Nuclear morphology and DNA fragmentation
The roots were fixed with 4% paraformaldehyde (w/v) and stained with 1 µg mL‑1 4′,6-diamidino-2-phenylindole (DAPI) for 30 min in the dark (Schweizer 1976). Fifty nuclei were counted and expressed as the number of nuclear deformations per 100 cells for all treated groups and expressed as a percentage. DNA fragmentation was analyzed using the TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) reaction and DNA gel electrophoresis (Hameed et al. 2004). TUNEL reaction was obtained with the Sigma-Aldrich ApopTagR Plus Fluorescein In situ Apoptosis Detection kit (Merck KGaA, Darmstadt, Germany). Light and fluorescence microscopic results were obtained using KAMERAM software, assisted by a KAMERAM camera and an Olympus BX-51 integrated microscope (Argenit, Istanbul, Türkiye). For DNA gel electrophoresis, genomic DNA was isolated according to Hameed et al. (2004). DNA isolates (100 ng μL-1) were run on a 0.8% (w/v) agarose gel at 100 V for 2 h. Gel was analyzed using the Isogen ProXima (2850) transilluminator and photographed using ProXima AQ-4 software (LabExchange, Germany).
Isolation of mitochondria and measurement of mitochondria inner membrane potential (ΔΨm)
Fresh roots weighing 1 g were ground in 2 mL of cold homogenization buffer (0.4 M sucrose, 50 mM Tris-HCl pH 7.4, 1 mM ethylenediaminetetraacetic acid (EDTA)). Filtered homogenates were then centrifuged at 1500 ×g for 15 min at 4 °C. The supernatants were used as cytosolic fractions, while pellets were resuspended in mitochondrial suspension buffer (0.4 M sucrose, 50 mM Tris-HCl pH 7.4), centrifuged at 14000 ×g for 15 min, and used as a mitochondrial fraction (Panda et al. 2008). Isolated mitochondria were diluted in a buffer (220 mM sucrose, 68 mM mannitol, 10 mM KCl, 5 mM KH2PO4, 2 mM MgCl2, 500 μM ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA), 5 mM succinate, 2 μM rotenone, 10 mM 4-(2-Hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES), pH 7.2) to a concentration of 0.1 mg mL-1, then rhodamine-123 was added (10 μg mL-1) and incubation in the dark followed for 5 minutes. The BiotekCytation microplate reader (Agilent, US) was used to measure ΔΨm-dependent quenching of Rh-123 fluorescence at excitation 490 nm and emission 535 nm (Huang et al. 2014).
Detection of cytochrome c release
Proteins (20 μg mL-1) from cytoplasmic and mitochondrial fractions were separated with 12% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes using transfer buffer (0.29 g glycine, 0.58 g Tris, 0.037 g sodium dodecyl sulfate (SDS), 20 mL methanol). The membrane was then blocked with 5% (w/v) skim milk solubilized in buffer (8 g NaCl, 0.2 g KCl, 3 g SDS, pH 7.4) at +4 ºC overnight. Subsequently, the membrane was cut into two parts between 35 and 25 kDa according to the protein marker. The upper part of the membrane was probed with the monoclonal antibody against mouse ATP5A (1:1000 dilution with 5% Tween 20 in PBS, Novusbio NBP2-15512) as mitochondrial marker protein (housekeeping protein). The lower part of the membrane was incubated with the monoclonal antibody against mouse cyt-c (1:1000 dilution with 5% Tween 20 in PBS, Novusbio NB100-56503). Goat anti-mouse IgG horseradish peroxidase (HRP)-conjugated secondary antibody was added at a dilution of 1:10000 with 5% Tween 20 in PBS. Membranes were stained with 0.06% (w/v) 3,3′-diaminobenzidine (DAB). Membranes were analyzed using the Isogen ProXima (2850) transilluminator and photographed using ProXima AQ-4 software (LabExchange, Germany), and the relative density of the bands was determined with the Image J program. The protein bands of cyt-c (in both cytoplasmic and mitochondrial fractions) were normalized by the ATP5A protein marker bands (housekeeping protein) using the formula:
Relative band density = density of cyt-c of sample / density of ATP5A
Determination of caspase-1-like activity
Caspase-1-like activity was measured according to the Caspase 1/ICE Colorimetric Assay Kit (ENZO Life Sciences, Switzerland). The roots (0.5 g) were ground in 1 mL cold extraction buffer (50 mM HEPES-KOH pH 7, 10% sucrose, 0.01% Chaps (3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate), 5 mM dithiothreitol, 1 mM EDTA) (Lombardi et al. 2007). The homogenate was centrifuged at 14000 ×g for 10 min at 4 °C. Supernatants were assayed for caspase-1-like activity, using YVAD pNA (N-acetyl-L-tyrosyl-L-valyl-L-alanyl-N-(4-nitrophenyl)-L-α -asparagine) as substrate. After incubation at 37 °C for 1 h, the absorbance was measured at 405 nm. Caspase-1-like activity was calculated according to the standard curve (y = 0.0435×), and expressed as nmol L-1.
RNA isolation, cDNA synthesis, and expression analysis of TaVPE4
Fresh roots weighing 100 mg were collected into RNase-free tubes and homogenized in liquid nitrogen. Total RNA was extracted using the Qiagen RNeasy Plant Mini Kit (Catalog No./ID: 74904), following the manufacturer's instructions. Next, cDNA was synthesized from the total RNA using QuantiTect Reverse Transcription Kit (Catalog No./ID: 205311). The expression of TaVPE4 was analyzed by semi-quantitative RT-PCR using Applied Biosystems Real-Time PCR (USA). The specific primers for semi-quantitative RT-PCR experiments were obtained from Kang et al. (2013). The actin gene was used as an internal control. PCR was performed using the cDNA templates and a Sigma-Aldrich PCR core kit with Taq DNA polymerase (CORET-1KT). The PCR products were loaded onto 1% agarose gels stained with 2 μL RedSafe nucleic acid staining solution (20.000×, Intron 21141) in Tris/Borat/EDTA buffer.
Quantitative PCR expression analysis of TaMCA1 and TaMCA4
The expressions of TaMCA1 and TaMCA4 genes were analyzed using quantitative real-time PCR (qPCR) using synthesized cDNA templates and specific primers [TaMC1-F GCGGATACTTCAGCCTTGTC (NCBI accession no KU958719.1); TaMC1-R CTTCCCGTGTTCCGTATTGT; TaMC4-F CCTCAAAGAGACCGTTCGTG (NCBI accession no JN807891.1); TaMC4-R ATCCTTCCCAGTTTGCTCCT]. The qPCR was performed on Applied Biosystems StepOneTM Real-Time PCR (USA) using Thermo Fisher reagents. Relative gene expression levels were calculated using the 2-ΔΔCt formula, with the TEF1 gene serving as an internal control (Livak and Schmittgen 2001).
Statistical analyses
All analyses were conducted with three biological and three technical replicates. Statistical analysis was performed using SPSS statistics 21.0. Significant differences were determined using one-way analysis of variance (ANOVA) followed by Tukey’s posthoc HSD test (P < 0.05).
Results
The mitotic index (MI) was 11.78% in control, 9.01% in 0.5 mg L-1, 8.13% in 1 mg L-1, 7.04% in 5 mg L-1, 6.88% in 10 mg L-1, and 5.51% in 20 mg L-1 of AgNP. The dose-dependent reduction of MI was observed after AgNP treatments, statistically significant, however, only at 20 mg L-1 (Fig. 1a). The percentage of chromosomal abnormalities (CA) induced by AgNPs in wheat roots was found to increase by 3.6, 4.5, 5.2, 5.3, and 5.5 fold with 0.5, 1, 5, 10, and 20 mg L-1 of AgNP, respectively (Fig. 1b).
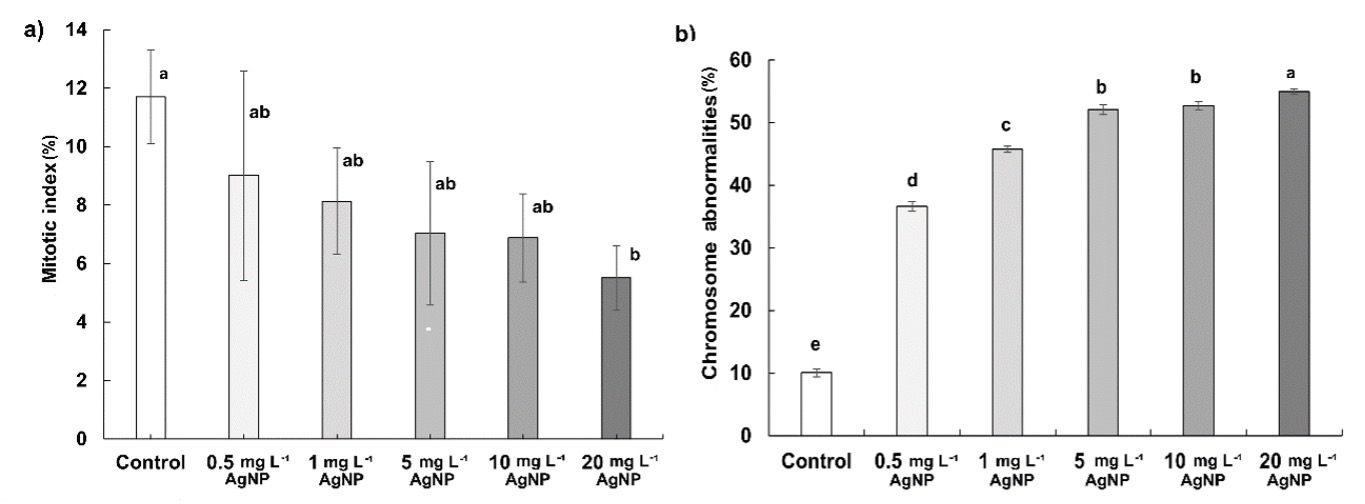
Fig. 1. Effects of 0.5, 1, 5, 10, and 20 mg L-1 of AgNPs on the mitotic index (a) and chromosome abnormalities (b) in wheat roots after 15 days of exposure. Data with different letters are statistically significant (P < 0.05) based on Tukey's HSD test. Bars on columns represent means ± standard deviation.
The frequency of cells with CAs (c-mitosis, anaphase bridges, multiple chromosome bridges, sticky chromosome, unoriented metaphase) increased in a dose-dependent manner in comparison to control. Semi-thin sections of control and treated wheat roots revealed that AgNPs affected the root diameter (On-line Suppl. Fig. 1.). Roots treated with AgNPs exhibited anatomical changes, including shrinkage of epidermal and cortical cells and a decrease in cell diameter. In roots treated with 0.5, 1, and 5 mg L-1 of AgNPs, deformation was observed in the epidermal and cortical cells, while no deformation was observed in the endodermis and central cylinder. Relatively high degeneration in root anatomy was observed in samples treated with 10 and 20 mg L-1 of AgNP.
The samples were stained with DNA-specific fluorescent dye DAPI to detect alterations in nuclear morphology. DAPI staining showed loss of nucleus spherical shape and exhibited chromatin condensation in wheat roots exposed to 0.5 mg L-1 of AgNPs. Similarly, nuclear deformation gradually increased with the increase of AgNP dose; it was 2% in control, 10% in 0.5 mg L-1, 12% in 1 mg L-1, 14% in 5 mg L-1, 20% in 10 mg L-1, and 24% in 20 mg L-1 of AgNP (Fig. 2).
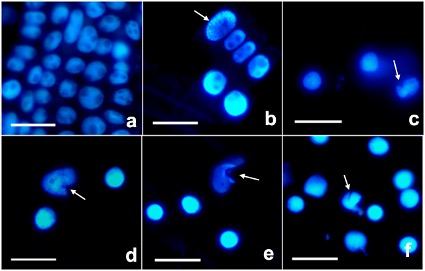
Fig. 2. Nucleus morphology in control wheat root cells ( a) and after 15 days of exposure to 0.5 ( b), 1 ( c), 5 ( d), 10 ( e), and 20 mg L-1 of AgNPs ( f). Arrows indicate nuclear deformation. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Scale bar = 50 μm.
The TUNEL reaction was used to detect the presence of free 3′ OH in DNA strand breaks. No TUNEL-positive nucleus was observed in the control, while a low number of TUNEL-positive nuclei was visible in all AgNP-treated roots (Fig. 3).
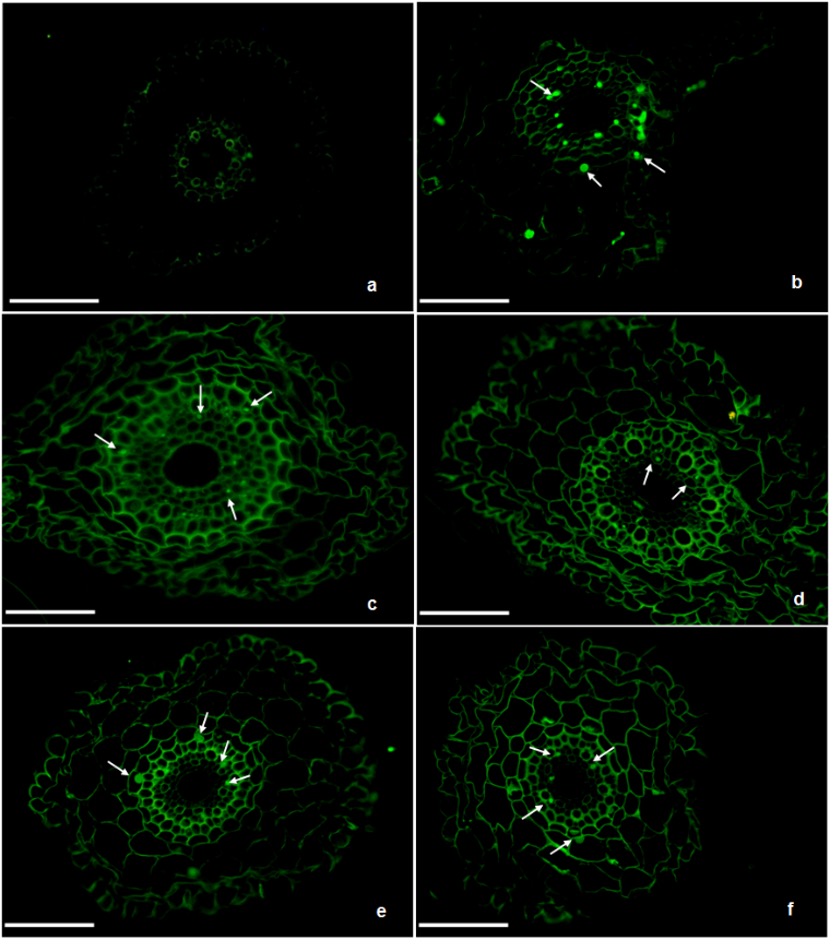
Fig. 3. TUNEL reaction in control wheat root cells ( a) and after 15 days of exposure to 0.5 ( b), 1 ( c), 5 ( d), 10 ( e), and 20 mg L-1 of AgNPs ( f). Images were obtained by TUNEL staining. Arrows indicate TUNEL-positive nuclei. Scale bars = 100 μm.
Our gel electrophoresis results revealed a slight induction of DNA smearing with no sign of DNA laddering, which was observed only in samples treated with 10 and 20 mg L-1 of AgNP concentrations (Fig. 4).
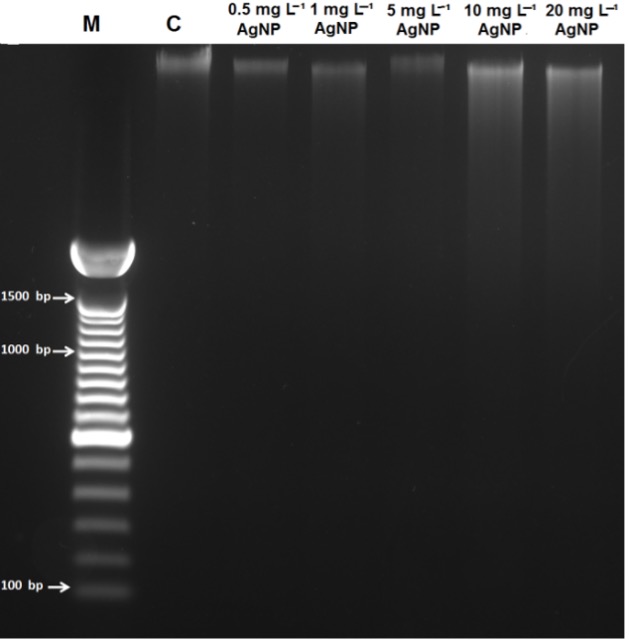
Fig. 4. DNA gel electrophoresis of control wheat root cells ( C) and after 15 days of exposure to 0.5 mg L-1, 1 mg L-1, 5 mg L-1, 10 mg L-1, and 20 mg L-1 of AgNPs. M = 1 kb DNA marker.
The early indications of PCD include alterations in mitochondrial morphology, loss of internal membrane potential, and cytochrome c (cyt-c) release from mitochondria to cytosol. According to our results, mitochondrial inner membrane potentials decreased significantly below the value obtained for control in a dose-dependent manner (Fig. 5a). The reduction was 7% at 0.5 mg L-1, 11% at 1 mg L-1, 15% at 5 mg L-1, 22% at 10 mg L-1, and 30% at 20 mg L-1 of AgNP in comparison to control. Furthermore, western blot analysis revealed the release of cyt-c to cytosol in all AgNP-treated roots (Fig 5c). However, we observed no significant differences in the cyt-c release between treatments and control based on the relative density of bands (Fig. 5b).
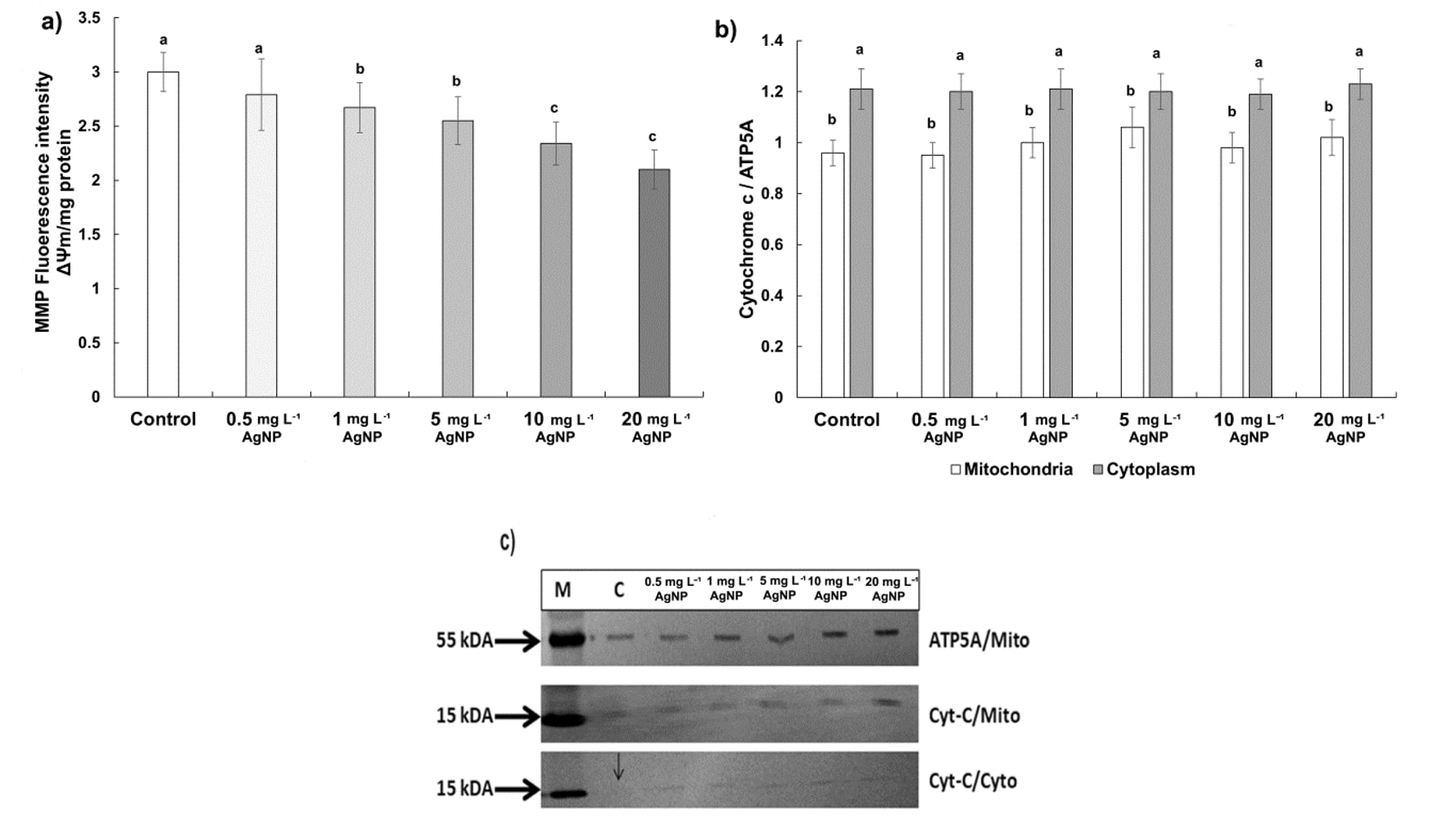
Fig. 5. Mitochondria inner membrane potential and cytochrome c expression in control wheat root cells and after 15 days of exposure to 0.5, 1, 5, 10, and 20 mg L-1 of AgNPs. Mitochondrial membrane potential (MMP, ΔΨm) ( a), comparison of relative density of cytochrome c (cyt-c) in mitochondrial and cytosolic fractions ( b), western blot analysis of cyt-c in mitochondrial and cytosolic fractions ( c). Data with different letters are statistically significant (P < 0.05) based on Tukey's HSD test. Bars on columns represent means ± standard deviation. Immunodetection was performed on three biological replicates, and one representative membrane was selected for display. ATP5A: marker protein, Cyto: Cytoplasmic fraction, C: Control, M: Marker, Mito: Mitochondrial fraction.
We observed a significant increase in caspase 1-like activity, by 9%, 33%, 13%, 43%, and 49% at concentrations of 0.5, 1, 5, 10, and 20 mg L-1 concentrations of AgNP treatments, respectively (Fig. 6).
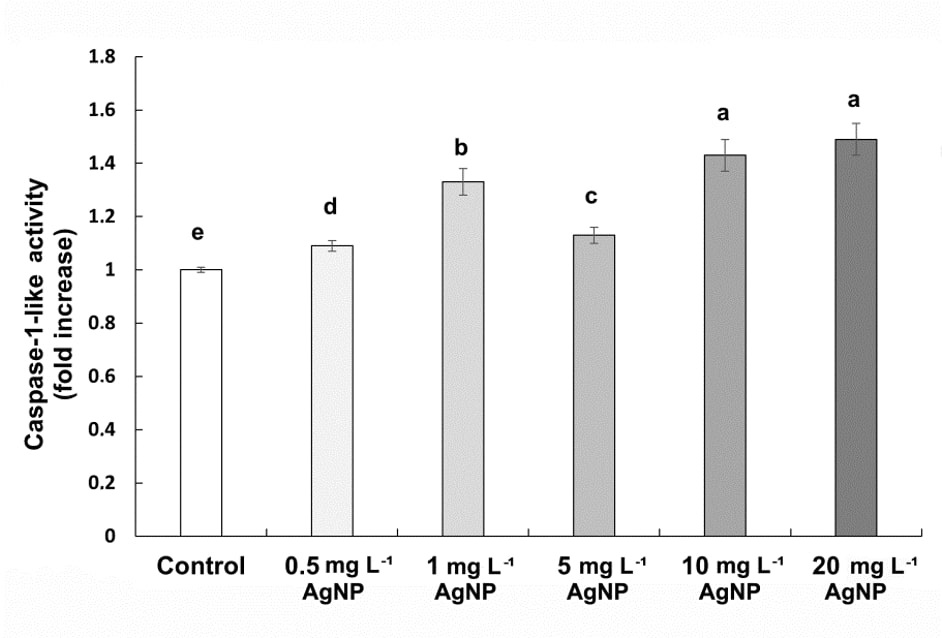
Fig. 6. Caspase-1-like activity in control wheat root cells and after 15 days of exposure to 0.5, 1, 5, 10, and 20 mg L-1 of AgNPs. Data with different letters are statistically significant (P < 0.05) based on Tukey's HSD test. Bars on columns represent means ± SD.
Furthermore, according to PCR results, TaVPE4 gene expression increased significantly at all applied concentrations of AgNP compared to the control (Fig. 7). The relative band density increased by 45% at 0.5 mg L-1, 139% at 1 mg L-1, 81% at 5 mg L-1, 80% at 10 mg L-1, and 145% at 20 mg L-1 of AgNP treatments compared to control. The increase in expression was particularly notable at concentrations of 1 and 20 mg L-1 of AgNP.
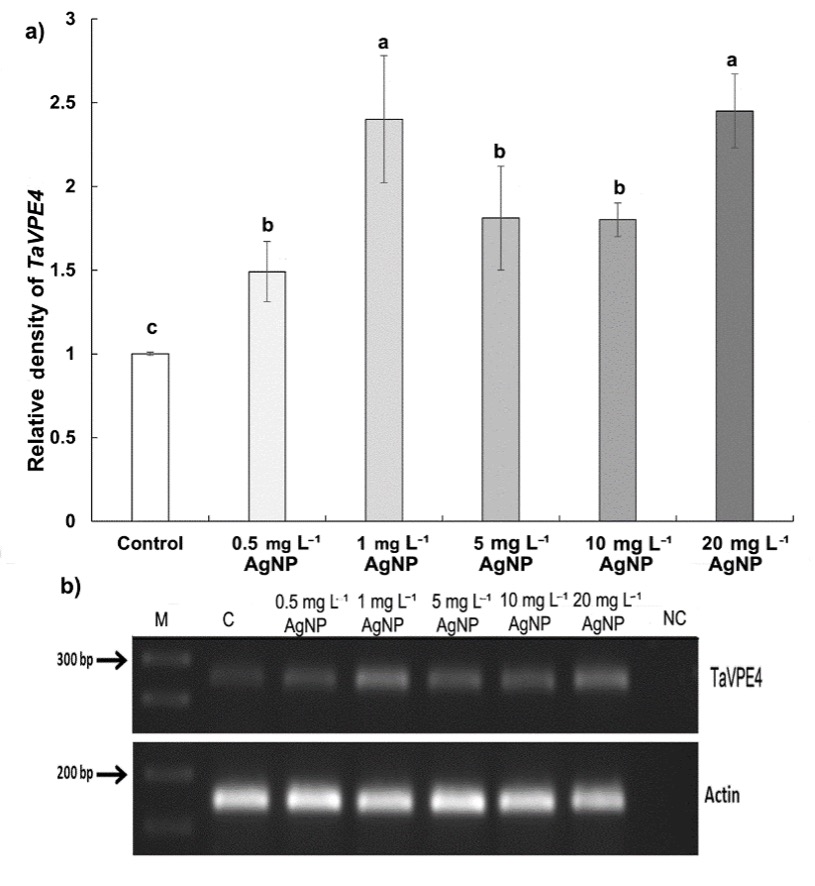
Fig. 7. Expression of TaVPE4 gene in control wheat root cells and after 15 days of exposure to 0.5, 1, 5, 10, and 20 mg L-1 AgNPs. Relative density of TaVPE4 (a), band intensities acquired by semi-quantitative PCR (arrows indicate 300 bp and 200 bp marker DNA) (b). Data with different letters are statistically significant (P < 0.05) based on Tukey's HSD test. Bars on columns represent means ± standard deviation. Expression analyses were performed on three biological replicates, and one representative gel was selected for display.
According to qPCR results, TaMCA1 and TaMCA4 exhibited differential expression patterns across AgNP concentrations. TaMCA1 was significantly up-regulated at 10 and 20 mg L-1 of AgNP, while no statistically significant difference was observed in the 0.5, 1, and 5 mg L-1 of AgNP treatments. In contrast, TaMCA4 was up-regulated at 1 and 5 mg L-1 of AgNP, whereas no statistically significant change was observed at 0.5, 10, and 20 mg L-1 of AgNP (Fig. 8).
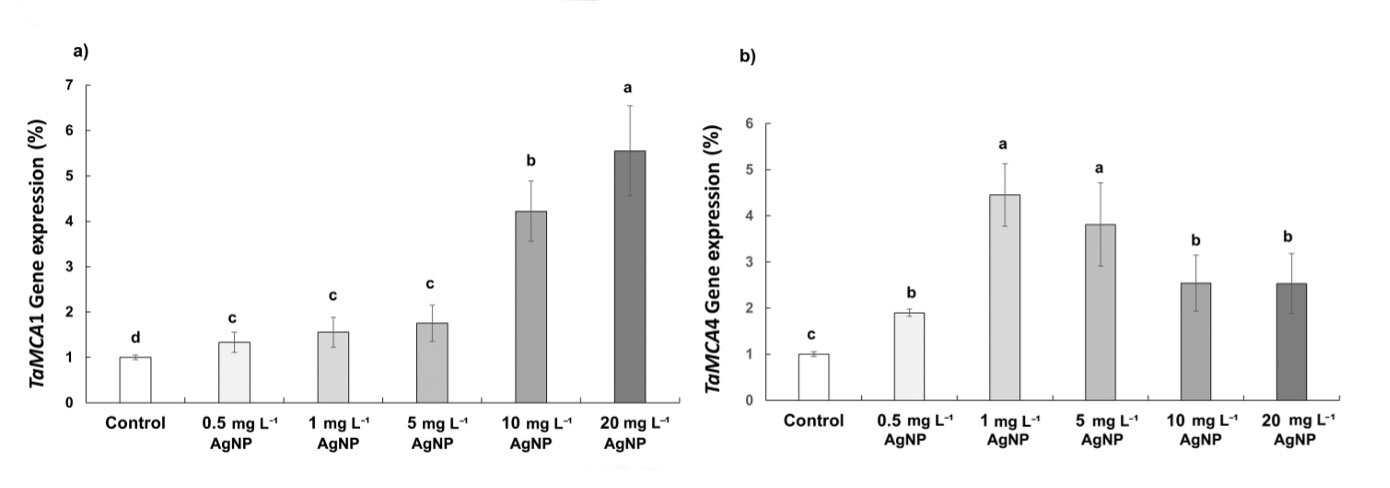
Fig. 8 The expression of TaMCA1 ( a) and TaMCA4 genes ( b) in control wheat root cells and after 15 days of exposure to 0.5, 1, 5, 10, and 20 mg L-1 of AgNPs. Data with different letters are statistically significant (P < 0.05) based on Tukey's HSD test. Bars on columns represent means ± SD.
Discussion
AgNPs are widely utilized daily and across various industries. It is estimated that European countries produce 5.5 tons of AgNP annually, increasing their content in the soil by 4 mg L-1 at a rate of 1.2 μg L-1. The US produces between 2.8 and 20 tons of AgNPs yearly, leading to an annual soil content increase of 6.6 μg L-1 to 2.3 mg L-1. On a global scale, the yearly input of AgNPs into soil ranges from 15 to 77 tons, with production expected to reach 400-800 tons by 2025 (Pulit-Prociak and Banach 2016, Kulikova 2021). Current estimations suggest that AgNP content in sewage sludge varies from 1.31-4.44 mg L-1 in Europe and 1.29-5.86 mg L-1 in the US. Some studies indicate that the highest safe AgNP concentration in soil is 0.05 mg L-1, and other estimates predict this concentration could be reached within 50 years in some European countries (Schlich et al. 2013, Kampe et al. 2018). Considering these accumulation values and estimations, we investigated cell death features after AgNP treatments which correspond to 0.5, 1, 5, 10, and 20 mg L-1 AgNP.
The percentage of cells in mitosis reveals the rate of cell division and low MI can be correlated with growth regression, genotoxicity, and cell death (Yanık et al. 2017). Consistently with the reduced MI and elevated CA results in our study, several researchers have reported an AgNP-induced increase in the frequency of CA and a decrease in MI in various species, including Allium cepa (Fouad and Hafez 2018), Drima polyantha (Daphedar and Taranath 2018), and T. aestivum (Abdelsalam et al. 2018). Furthermore, AgNP treatments adversely affect the root anatomy of wheat (Abdelsalam et al. 2018). Mirzajani et al. (2014) stated that AgNPs permeated the cell wall and damaged the cell morphology and structure of Oryza sativa roots. Similarly, Tripathi et al. (2017) observed irregularities in the shape of cortical cells on cucumber roots following AgNP exposure. Cvjetko et al. (2018) revealed that tobacco root cells are highly vacuolated and absorb AgNPs in the meristem cells, indicating vacuolization where AgNPs are stored. Likewise, our previous study confirmed the uptake of AgNPs by roots, their accumulation in the cytoplasm of cortical cells, and significant inhibition of root elongation in higher concentrations (10 and 20 mg L-1) (Yanık and Vardar 2019). When our previous (Yanık and Vardar 2019) and present results are evaluated together, it can be concluded that regression in root elongation may be related to the decrease in MI and increase in CA due to AgNP accumulation and toxicity.
Chromatin condensation, its passage through the nucleus periphery, and degradation of nuclear DNA are among the most prominent characteristics observed in cells undergoing PCD. During chromatin condensation, DNA is firstly cleaved into high molecular weight fragments (50-300 kb), followed by low molecular weight fragments (Minina et al. 2021). We observed alterations in nuclear morphology, chromatin condensation, and slight DNA smear, particularly in response to higher AgNP concentrations (10 and 20 mg L-1). Several researches have shown that aluminum (Al) nanoparticles (AlNPs) induce chromatin condensation and DNA fragmentation in T. aestivum roots, as evidenced by DAPI staining, TUNEL assay, and gel electrophoresis (Yanık and Vardar 2015, Yanık et al. 2017). Additionally, Thiruvengadam et al. (2015) found that different concentrations of AgNPs (1, 5, and 10 mg L-1) cause DNA damage in Brassica rapa roots, as confirmed by DNA laddering, comet assay, and TUNEL staining. These results show that both NPs and AgNPs can induce chromatin and/or DNA damage in a concentration-dependent manner, which is consistent with the observations in our experiments.
The earliest characteristic indications of PCD in plants include alterations in mitochondrial morphology, loss of inner mitochondrial membrane potential, and cyt-c release to the cytosol (Paes de Melo et al. 2022). Cyt-c is localized in the mitochondrial intermembrane space and is linked to the inner mitochondrial membrane. Under stress conditions, ROS regulates cardiolipin oxidation, resulting in cyt-c diffusion into the intramembrane region. Additionally, ROS triggers the mobilization of Ca2+ ions from the endoplasmic reticulum (ER) lumen to the mitochondria. These Ca2+ ions can induce a decrease in mitochondrial transmembrane potential and the opening of mitochondrial permeability transition pores (MPTP) in the outer mitochondrial membrane, facilitating the cyt-c release to the cytoplasm (Huang et al. 2014, Paes de Melo et al. 2022). The release of cyt-c further stimulates ROS generation within the mitochondria, creating a positive feedback loop to enhance the initial signal. ROS production and cyt-c release may be prerequisites for the PCD execution rather than directly activating it (Reape and McCabe 2008). Previous studies have observed cyt-c release in plants under various stress conditions, such as heavy metal exposure, temperature, fusarium toxin, salt, and flooding (Li et al. 2007, Sarkar and Gladish 2012, Huang et al. 2014). For example, Huang et al. (2014) investigated Al toxicity in the roots of Arachis hypogaea and found that it induces DNA fragmentation, chromatin condensation, caspase-3-like activity, and reduction in mitochondrial inner membrane potential and membrane permeability, resulting in the cyt-c release from the mitochondria to the cytoplasm. More recently, Qi et al. (2018) showed that waterlogging stress led to the degradation of mitochondrial structure, increase of the mitochondrial permeability transition, and decrease of mitochondrial transmembrane potential and cyt-c release in the endosperm of wheat as evidence of PCD process. In the presented study similarly, a significant decrease of mitochondrial inner membrane potentials and release of cyt-c to cytosol in all AgNP-treated wheat roots were observed, for the first time as far as we know.
PCD is regulated by proteases belonging to five distinct categories: serine, aspartate, cysteine, threonine, and metalloproteases (Rawlings et al. 2018). While the most well-known proteases involved in the initiation and execution of animal apoptosis are caspases, which cleave their substrates after an aspartate residue using a catalytic dyad consisting of cysteine and histidine amino acids (Klemenčič and Funk 2018), plants lack true caspase. Instead, plant PCD is thought to be mediated by caspase-like or other protease enzymes (Salguero-Linares and Coll 2019). Among the main cysteine proteases in plants are vacuolar processing enzymes (VPEs), metacaspases (MCs), and papain-like cysteine proteases (PLCPs). These proteases play crucial roles in various physiological processes, including PCD (Salguero-Linares and Coll 2019). Hatsugai et al. (2015) showed that VPEs have structural and enzymatic properties similar to mammalian caspase-1, displaying YVADase/caspase-1-like cleavage activity despite limited sequence identity to caspase. It has also been revealed that the synthetic caspase-1 inhibitor Ac-YVAD-CHO inhibits the activation of VPE (Hatsugai et al. 2015). Various studies focusing on PCD have examined VPE gene expression and caspase-like activity. For example, Kariya et al. (2013) reported that the VPE activity increases with the loss of plasma membrane integrity in cell lines exposed to 100 μM Al, and caspase-1 inhibitor suppresses PCD in Nicotiana tabacum cell line BY-2. It was stated that the expression of VPE genes was significantly enhanced by Al treatment. Moreover, many studies have demonstrated that various abiotic stressors, such as ethylene, salicylic acid, gibberellic acid, nitric oxide, H2O2, and salt, upregulate VPE genes across different plant species (Kang et al. 2013, Wang et al. 2018). Kang et al. (2013) reported temporal and spatial expression patterns of TaVPEs in different tissues of wheat under various stress conditions. After treatment with NaCl and H2O2, TaVPEs were upregulated in roots, particularly TaVPE4, which showed extensive expression. After treatment with NaCl, the expression of TaVPE4 was remarkably high for 24 hours, but then it decreased. On the other hand, the expression of TaVPE4 was significantly high in roots after H2O2 treatment, and it remained consistent over time. NaCl treatment slightly induced TaVPE1 and TaVPE2 in roots. After 48 hours of treatment, visible transcripts of TaVPE1 and TaVPE2 were detected in leaves (Kang et al., 2013). Based on this study, we observed that TaVPE4, which is expressed in roots, especially under stress conditions, was significantly upregulated compared to the control and reached the highest value, especially at 1 and 20 mg L-1. To the best of our knowledge, our study is the first to demonstrate the induction of TaVPE4 gene expression and caspase 1-like activity by AgNP treatment.
Metacaspases represent the largest subfamily of cysteine proteases and are found in plants, fungi, and protists based on their caspase homology. They are classified into three categories – type I, type II, and type III, based on the architecture of their p10 and p20 domains. Unlike caspases, metacaspases exhibit substrate specificity to arginine or lysine instead of aspartate. Several reviews have compiled studies suggesting that plant metacaspases are involved in dPCD and ePCD (Minina et al. 2017, 2021). TaMCA1 is a type I metacaspase and it has been reported that while the TaMCA1 gene does not exhibit caspase-1-like activity, it could suppress PCD in tobacco and wheat leaves induced by the mouse Bax gene in interaction with pathogen Puccinia striiformis (Pst) (Hao et al. 2016). Hao et al. (2016) also indicated that the knockdown of the TaMCA1 gene increases plant resistance to Pst. They reported that the expression level of TaMCA1 increased significantly at 48 and 72 hours post inoculation (hpi) with Pst, indicating that TaMCA1 may play an important role in the interaction between wheat and Pst. Their research suggests that TaMCA1 regulates cell death only after generating sufficient signals, such as mammalian Bax, Pst, or H2O2. Our results showed that TaMCA1 up-regulated significantly only in 10 and 20 mg L-1 of AgNP treatment. It may be increased to suppress PCD in these stages where only DNA fragmentation is observed.
Another wheat metacaspase, TaMCA4, is classified as a type II metacaspase and was identified by Wang et al. (2012). It was demonstrated that overexpression of TaMCA4 enhances PCD induced by Pst, while knockdown of TaMCA4 has the opposite effect, suggesting that TaMCA4 acts as a positive regulator of PCD (Wang et al. 2012). To assess the involvement of metacaspase genes in AgNP-induced PCD, we examined the expression of TaMCA1 and TaMCA4 genes. While TaMCA1 expression increased at 10 and 20 mg L-1, TaMCA4 expression up-regulated significantly only at 1 and 5 mg L-1 AgNP treatment. Based on these results, we speculate that TaMCA1 and TaMCA4 genes may work antagonistically and they may suppress each other. Moreover, different concentrations of AgNPs may induce the activity of different types of metacaspases to regulate ePCD.
While numerous studies have investigated the accumulation, translocation, and toxicity of NPs in plants, the phytotoxicity mechanisms remain unclear. Additionally, the execution and regulation of NP-induced PCD are still not fully understood. This study is the first to examine AgNP-induced PCD in wheat root cells. Our results showed that AgNPs at different concentrations (0.5, 1, 5, 10, and 20 mg L-1) induce PCD to varying degrees. This research contributes to our understanding of NPs’ role in regulating and executing PCD, which is crucial for advancing knowledge on the long-term effects of NPs on ecosystems and crop yield stability in current and future investigations.
