Introduction
Nearly 200 types of cheese are produced in Türkiye, which vary based on different milk types, production techniques, ripening times, and conditions (Hacıoğlu and Kunduhoğlu, 2021). Tulum cheese is produced in many regions of Türkiye and is the most consumed cheese type, along with White and Kaşar cheese. It is characteristically white or cream, high in dry matter and fat, and has a distinctive aroma, homogeneous texture, and pronounced acidic flavour (Oluk et al., 2014).
Among Türkiye's artisanal cheeses, Muş Tulum cheese stands out for its consumer appeal. This artisanal semi-hard cheese is produced exclusively from unpasteurized sheep milk in the Eastern Anatolian region of Türkiye from May to August, without using any starter cultures.
Various enzymes and indigenous lactic acid bacteria (LAB) present in milk are crucial in improving cheese quality. Through microbiological and biochemical changes, LAB contributes to developing a complex flavour and aroma profile (Silva et al., 2015). LAB also generate various antimicrobial substances, including bacteriocins, hydrogen peroxide, carbon dioxide, diacetyl, ethanol, and organic acids. These compounds inhibit the growth of undesirable contaminants, thus prolonging the shelf life of cheese (Agriopoulou et al., 2020; Montel et al., 2014).
The lactic microflora in cheese comprises two types of bacteria: starter lactic acid (SLAB) and non-starter lactic acid (NSLAB). SLABs are intentionally added to the milk and are responsible for producing acid during cheese production. On the other hand, NSLAB are naturally found in raw milk and play a significant role in the development of cheesy flavour (Psomas et al., 2023; Vandera et al., 2019).
Indigenous LAB is mainly involved in the formation of the characteristic flavour and aroma of traditional raw milk cheeses include species such as Lactobacillus spp. (Lb. plantarum, Lb. paraplantarum, Lb. caesi, Lb. paracasei, Lb. fermentum, Lb. brevis, Lb. pentosus, Lb. rhamnosus), Lactococcus spp. (Lc. lactis subsp. lactis, Lc. cremoris), Enterococcus spp. (E. faecium, E. feacalis, E. durans), Streptococcus spp. (Str. thermophilus), Leucocnostoc spp. (Leu. mesenteroides and Leu. pseudomesenteroides), Pediococcus spp. and Weisella spp. species (Vandera et al., 2019; Bluma et al., 2017; Milani et al., 2016; Gobbetti et al., 2015).
Pasteurization negatively affects the flavour quality of cheeses by inactivating various enzymes in milk, such as proteases and lipases, which are crucial for the formation of individual flavours and aromas in cheeses. In addition, pasteurization specifically inactivates the indigenous lactic microflora, which negatively affects the flavour profile (Tomasino et al., 2018; Jo et al., 2018; Kırmacı et al., 2016). While pasteurization of milk and use of commercial starter cultures eliminate safety risks related to consumer health, it is acknowledged that these processes do not fully capture the unique flavour profile produced by indigenous LAB in cheese (Psomas et al., 2023).
LAB strains isolated from raw milk cheeses enhance the development of a more complex and aromatic taste and scent than commercially produced starter cultures (Picon et al., 2019; Baruzzi et al., 2016). Various studies aiming to discover the indigenous microorganisms in cheeses produced without starter cultures are of increasing interest. The limited variety of starter cultures employed in the dairy industry and the rising consumer desire for more flavourful products have sparked a growing interest in utilizing indigenous LAB strains as starters (Uymaz et al., 2019; Bozoudi et al., 2016).
Considering the lack of previous research on the development of starter cultures to be used in the production of Muş Tulum cheese and the significant role of the indigenous lactic flora in terms of cheese quality as well as the search for new potential starter cultures; this study aimed to characterize the dominant lactic flora at strain level during storage in Muş Tulum cheese produced from raw sheep milk by biochemical, phenotypic and genotypic methods and to obtain data to determine the optimal starter culture type for use in industrial production.
Materials and methods
Tulum cheese samples
As shown in Figure 1, cheese samples produced in May-August 2022 in different geographical regions of Muş province were obtained from 5 different dairy factories (A, B, C, D, and E) and left to ripen for 3 months (2±1 °C, 80-85 % relative humidity). Samples were taken and analysed on the 1st, 45th, and 90th days of storage.
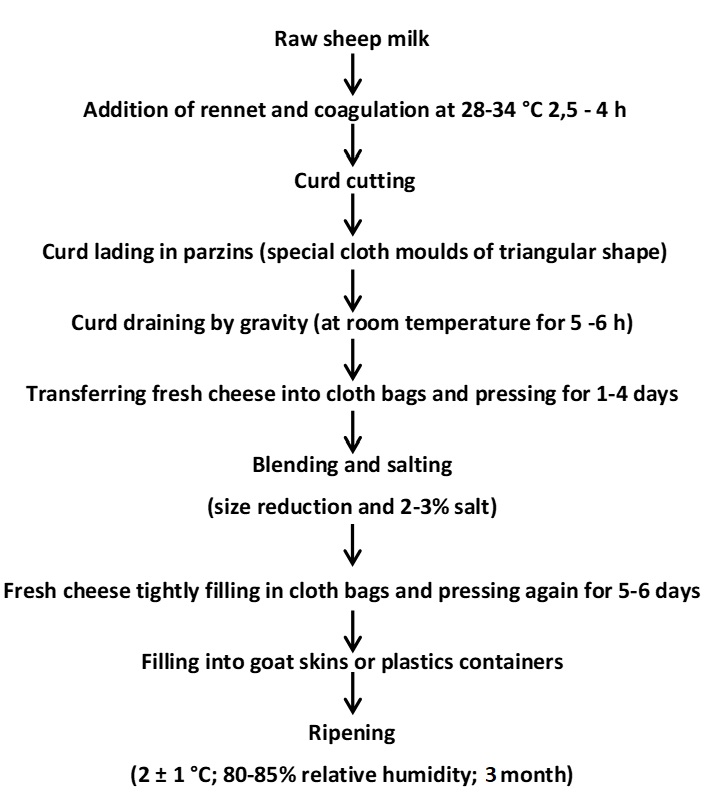
Figure 1. Traditional production of Muş Tulum cheese workflow
Enumeration and isolation of bacteria in cheese samples
The 20 g cheese samples were weighed and transferred to a stomacher (Colwarth Stomacher 400C Seward Laboratory, UK) with 180 ml of sodium citrate (Tekkim, Bursa, Türkiye) and then homogenized (Gerasi et al., 2003).
The three selective media used for enumerating and isolating different groups of bacteria are as follows: lactococci are grown on M17 agar at 30 °C for 48-72 hours under anaerobic conditions; lactobacilli are grown on MRS agar at 37 °C for 48-72 hours under anaerobic conditions; enterococci are grown on Slanetz Bartley (SB) agar at a temperature of 45 °C for 48 hours under anaerobic conditions. To achieve anaerobic conditions, anaerobic jars and anaerocult were used. The medium was procured by Merck (Darmstadt, Germany). Following the incubation period, three to four colonies were selected from the colonies developing in each petri dish and included in the study.
Identification of strains based on phenotype and biochemical properties
The phenotypic characterization of chosen 180 isolates from M17, MRS, and SB plates involved the performance of Gram staining, morphological analysis, and catalase testing. All purified isolates were maintained at -20 °C as frozen stocks containing 50 % glycerol (Sigma-Aldrich, Germany) until analysis. The growth of Gram-positive and catalase-negative isolates was assessed in a medium containing 4 %, 6.5 %, and 10 % NaCl and at temperatures of 10, 15, and 45 °C (Yüce, 2017). Furthermore, the gas generation resulting from glucose metabolism in the isolates was assessed using Durham tubes in MRS and M17 media supplemented with 2 % glucose (Fortina et al., 2003).
Genotypic characterization of strains by 16S rDNA sequence analysis
The process of extracting DNA isolation was performed using the methodology reported in a previous study (Osmanağaoğlu et al., 2010).
The study examined the intraspecific biodiversity of isolates from LAB using Randomly Amplified Polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR) analysis. A total of three to four colonies were selected from each petri dish. Considering the possibility that the selected colonies might be the same, RAPD analysis was performed at molecular level, and one of the strains with a similar band profile was selected and sent for sequence analysis. This research utilized two universal primers: OPA-7 (5'GAAACGGGTG 3') and OPA14 (5'TGCTGCAGGT 3'). After an initial denaturation at 94 °C for 1 minute, the products were amplified through 40 cycles. Each cycle involved denaturation at 94 °C for 1 minute, annealing at 36 °C for 1 minute, and elongation at 72 °C for 1 minute. The ultimate extension was conducted at 72 °C for 10 minutes. The PCR products were ultimately separated using 1 % (w/v) agarose gels that contained ethidium bromide. The gel was observed using a Kodak Gel Logic 200 Imaging device, an ultraviolet light bio-imaging device manufactured by KODAK in the United States.
Out of the identified isolates, 65 were used for sequencing the 16S rDNA gene after performing RAPD-PCR. The amplification was performed using the 20-F 5'-AGAGTTGATCCTGGCTCAG-3' and 1390-R 5' GACGGGCGGTGTGTACAA-3' universal primers. The PCR protocol was as follows: initial denaturation at 94 °C for 1 minute, followed by 30 cycles of amplification, each consisting of the following steps: denaturation at 94 °C for 1 minute, annealing at 54 °C for 15 seconds, and extension at 72 °C for 1 minute. Lastly, the final extension step was conducted at 72 °C for 10 minutes. The resulting PCR products were then sent to Genolysis Life Sciences and Technologies Joint Stock Company, located in the Agriculture, Livestock and Food Technopark in Ankara, Türkiye, for sequencing. The obtained sequences were compared to those in the NCBI GenBank database using the BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST).
Technological characterization of strains
Acidifying activity of isolates
The acid generation capacity of the isolates was assessed by measuring the pH and using the titrimetric method to calculate the percentage of lactic acid, following the protocol given by Sarantinopoulos et al. (2001a). The isolates were obtained from frozen stocks and grown in MRS and M17 medium. Lactococci were cultured at 30 °C, while enterococci and lactobacilli were cultured at 37 °C. The cultures were incubated for 24 hours. The obtained cultures were introduced into 10 mL of sterile UHT skim milk at a concentration of 1 % and kept in a controlled environment at a temperature of 37 °C. 2 mL sterile samples were collected at 3, 6, 9, and 24 hours during the incubation period. The pH was evaluated using an Orion model 250 A pH meter, and the titratable acidity was assessed.
Proteolytic activity of isolates
The isolate’s proteolytic activity was assessed using a modified version of the method of Saez et al. (2018). Samples were treated with 0.75 % trichloroacetic acid (1:3) (Biochem Chemopharma, Loire, France) and 150 µL of supernatant was deproteinized with 3 mL OPA reagent prepared by dissolving 40 mg OPA (Sigma Aldrich, USA) in 1 mL methanol, 25 mL 0.1M sodium tetraborate (Sigma-Aldrich, Germany), 2.5 mL of 20 % (m/V) sodium dodecyl sulfate (Sigma-Aldrich, USA) was mixed with 100 µL of β-mercaptoethanol (Merck, Damstadt, Germany) and the final amount was adjusted to 50 mL using distilled water (Sigma-Aldrich, Merck, USA) and the process took 10 minutes at room temperature. The absorbance measurement was performed at a wavelength of 340 nm using a Cary 60 UV-Vis spectrophotometer manufactured by Agilent Technologies in Santa Clara, CA, USA. The measurements were quantified in millimoles of free amino acids (FAA) per liter of milk. Proteolytic activity levels were evaluated as low (0-1 mM/L), medium (1-2 mM/L) and high (2-3 mM/L).
Bacteriocin activity of isolates
From cultures developed overnight at 37 °C, 2000 µL were withdrawn and placed into sterile microcentrifuge tubes. The tubes were then centrifuged at 12,000 rpm for 10 minutes. After centrifugation, 1000 µL of supernatant was aspirated with a micropipette, transferred to a sterile tube, and adjusted to a pH of 7.0 with 6 M NaOH. Subsequently, the samples were treated with catalase (1 mg/L) and filtered through 0.22 µm diameter filters. After filtration, the samples were boiled at 100 °C for 5 minutes. The indicator microorganism was Lactococcus lactis SIK 1403, cultured overnight at 37 °C. The study used the Pediococcus pentosaceous OZF strain, know to produce pediocin, as a control. The well diffusion method was performed according to the protocol established by Osmanağaoğlu et al. (2010).
Results and discussion
Changes in LAB numbers during the maturation of Tulum cheese
Table 1 displays the potential mean quantities of LAB throughout the maturation phase of Tulum cheese samples collected from several dairy farms. During the storage, the average colony numbers on MRS, M17, and SB plates ranged from 7.21 to 8.69, 7.73 to 8.71, and 7.00 to 7.76 log cfu/g, respectively. The findings align with the overall LAB counts (7.66-8.15 log cfu/g) seen in Muş Tulum cheese that underwent ripening using various packaging materials, as Rençber and Çelik (2021) documented. Nevertheless, they surpass the mean LAB counts documented in previous investigations, including those of Aktaş and Erdoğan (2022), Mohammed and Çon (2021), Evren and Şıvgın (2021), and Kara and Akkaya (2015).
Table 1. Changes in LAB numbers during the ripening period in Muş Tulum cheese (log cfu/g)
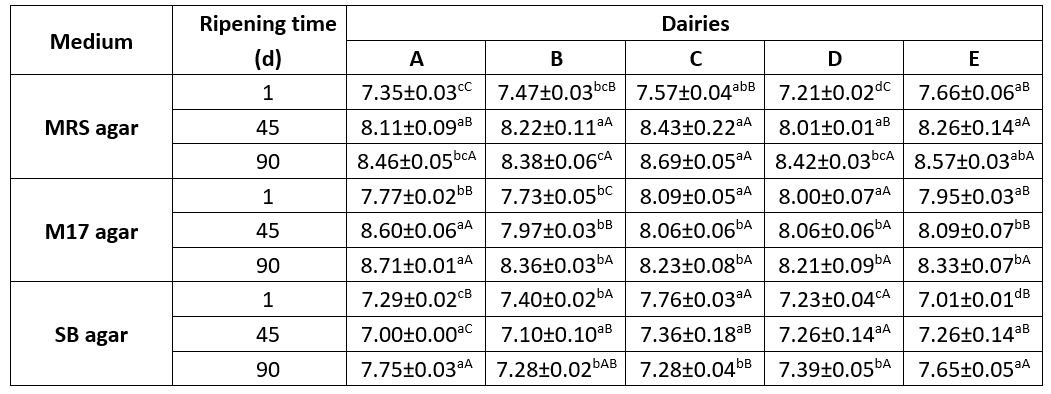
Values are the mean ± standard deviation; different uppercase and lowercase superscript letters indicate significant differences for the same sample within different days of ripening and between samples on the same ripening day, respectively (p<0.05)
The colony counts on MRS plates increased consistently during the storage period in all samples. This increase was considered to be significant at a p˂0.05 level in samples A and D throughout the storage period and in samples B, C, and E on the 45th day of storage. Similarly, colony counts on M17 plates increased consistently throughout all stages except for sample C. This increase was significant at a p˂0.05 level in sample B. The number of colonies on SB plates decreased on the 45th day of storage in samples A and B but then increased towards the end of the storage period. This change was significant at a p˂0.05 level in sample A. On contrary, it increased continuously in samples D and E throughout the ripening period.
When the bacterial counts in different media on the same storage days were compared among the samples, significant differences were observed in bacterial counts in all samples on MRS plates and in samples B, C, and E on SB plates on the 1st day of storage (p ˂ 0.05). Additionally, on the 90th day of storage, significant differences in bacterial counts on MRS plates were observed between samples B, C and E (p˂0.05). On the other hand, on the 45th day of storage, the change in bacterial counts was not significant (p˃0.05) for any of the samples. Rençber and Çelik (2021) reported a highly significant difference (p˂0.01) in LAB counts among dairy factories producing mature Muş Tulum cheese.
LAB strains phenotypic and biochemical identification
Isolates were subjected to morphological examination under a microscope, gram-staining and catalase tests. A total of 171 strains were obtained, with 59 isolated from MRS agar, 53 from M17 agar, and 59 from SB agar.
The isolates were assessed for their capacity to thrive under varying temperatures (10 °C, 15 °C, and 45 °C) and salt concentrations (4 %, 6.5 %, and 10 %) as indicated in Table 2. Out of 50 strains of Enterococcus and 4 Lactococcus tested, 35 Enterococcus strains and 2 Lactococcus strains (FFH51 and FFH53) were able to grow at all three temperatures and at 10 % NaCl. Nevertheless, the 15 remaining strains of Enterococcus and Lactococcus (FFH48 and FFH54) exhibited growth independently at a temperature of 45 °C and with a NaCl concentration of 10 %. They did not demonstrate any growth at a temperature of 10 °C. Several studies have examined the growth of enterococci under different conditions. Uymaz et al. (2019) and Kırmacı et al. (2016) tested the growth of all enterococcal species at 10 °C and 45 °C, respecitevly, and in a solution containing 6.5 % NaCl. Ertürkmen and Öner (2015) found that many enterococci and lactococci strains isolated from white cheese did not grow at 10 °C. Our investigation discovered that lactococci exhibited growth in NaCl concentrations of 6.5 % and 10 %, which contradicts the data given by Silva et al. (2022). All Lactiplantibacillus isolates, on the other hand, showed a growth at 15 °C, 45 °C and 10 % NaCl. Aktaş and Erdoğan (2022) reported that the Lactiplantibacillus strains they isolated from white cheese exhibited weak or no growth at 45 °C and 10 % NaCl.
Table 2. Some phenotypic and biochemical properties of LAB isolated from Muş Tulum cheese
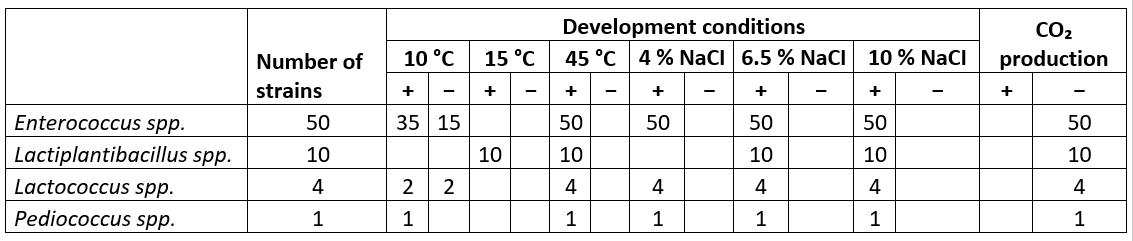
None of the isolated strains produced carbon dioxide from glucose, and all strains were determined to be homofermentative LAB (refer to Table 2). Several studies have found that all species of Enterococcus, Lactiplantibacillus, and Lactococcus are homofermentative (Aktaş and Erdoğan, 2022; Albayrak and Duran, 2021; Mohammed and Çon, 2021). These findings indicate that these strains have the potential to be employed as starter cultures in the production process of cheese.
Genotypic identification of LAB strains
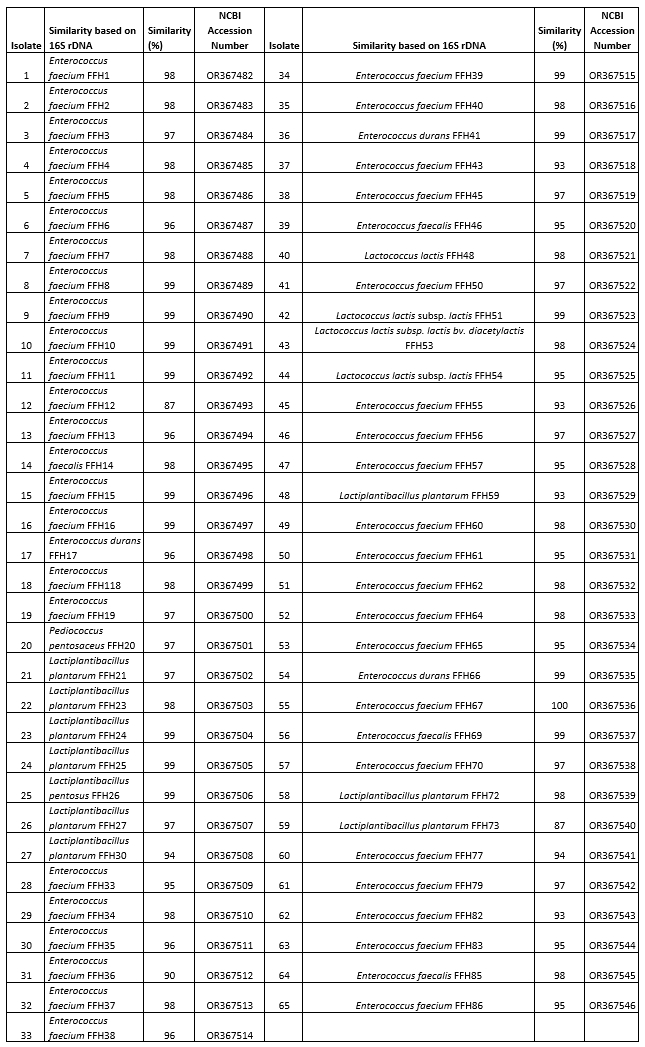
The isolates were genotypically characterized using 16S rDNA gene sequence analysis. The acquired sequences were compared to those in the NCBI GenBank database utilizing the BLAST program. After comparison, it was observed that the identified strains exhibited species-level similarities ranging from 87 % to 100 % (refer to Table 3). Based on the sequence analysis, the autochthonous LAB population was identified as Enterococcus spp. (76.92 %), Lactiplantibacillus spp. (15.38 %), Lactococcus spp. (6.15 %), and Pediococcus spp. (1.54 %).
Genetic analysis revealed that out of the 50 Enterococcus spp., 43 were classified as Enterococcus faecium (86 %), 4 as Enterococcus faecalis (8 %), and 3 as Enterococcus durans (6 %). Out of the 10 Lactiplantibacillus species, 9 were classified as Lactiplantibacillus plantarum, accounting for 90 % of the total, while 1 was identified as Lactiplantibacillus pentosus, making up 10 %. Four Lactococcus species were found, comprising of two Lactococcus lactis subsp. lactis (50 %), one Lactococcus lactis (25 %), and one Lactococcus lactis subsp. lactis biovar diacetylactis (25 %). In addition, one strain was identified as Pediococcus pentosaceus.
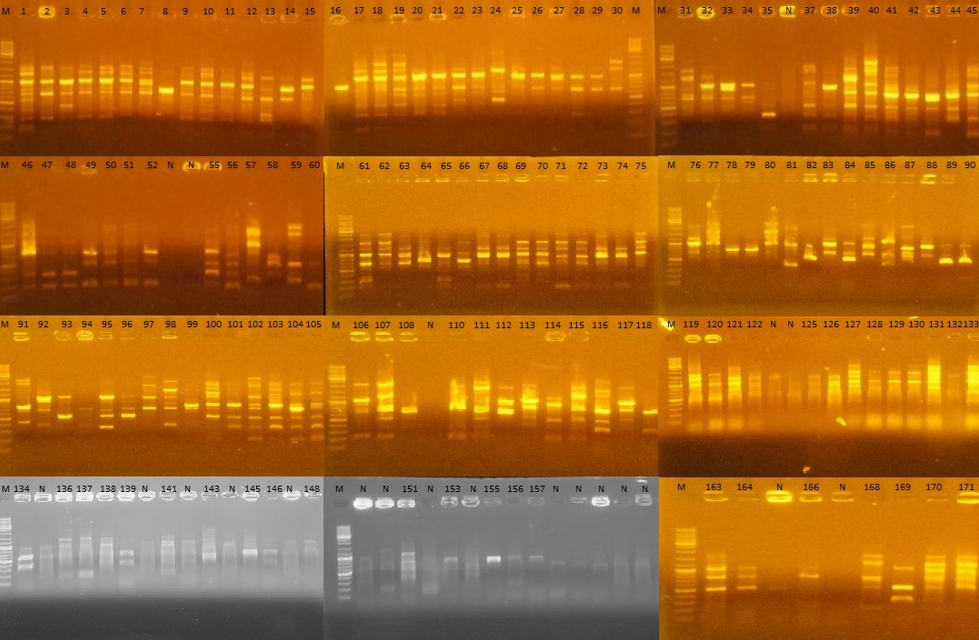
Figure 2. Randomly Amplified Polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR) gel images of 171 strains isolated from Muş Tulum cheese. M: Marker, N: Negative. 1-60; strains isolated on the 1st day of ripening, 61-118; strains isolated on the 45th day of ripening, 119-171; strains isolated on the 90th day of ripening
Out of the LAB strains sequenced, 27 (70.37 % Enterococcus spp., 25.92 % Lactiplantibacillus spp. and 3.70 % Pediococcus spp.) were isolated on the 1st day of ripening, 23 (78.26 % Enterococcus spp., 17.39 % Lactococcus spp. and 4.34 % Lactiplantibaciilus spp.) on the 45th day and 15 (86.66 % Enterococcus spp. and 13.33 % Lactiplantibacillus spp.) on the 90th day.
Table 3. Classification of bacterial groups according to subspecies and their degree of similarity as a result of 16S r DNA sequence analysis
The analysis of Muş Tulum cheese samples from 5 distinct dairy factories revealed that Enterococcus spp. was the dominant species during each stage of ripening, with Lactiplantibacillus spp. being the subsequent dominant species, except in one dairy factory (Lactococcus spp. was the second dominant species on the 45th day of ripening in sample D). Enterococcus spp. dominates at all stages of Muş Tulum cheese production because to its elevated tolerance to salt concentration and acidic conditions (Terzić-Vidojević et al., 2020; Elkenany et al., 2018). The presence of enterococci in Muş Tulum cheese may potentially be attributed to inadequate sanitation practices in the handling of raw milk or processing equipment. Enterococci are believed to be present in milk due to contamination from the animal's external surface, unhygienic milking equipment, milk storage tanks, or water sources that have been contaminated with feces (Kırmacı et al., 2016). These bacteria are important for the flavour development of certain cheeses, including Feta, Mozzarella, Cebreiro, and Venaco (Hayaloğlu, 2016). Furthermore, several strains of Enterococcus generate antimicrobial peptides that hinder the proliferation of unwanted microorganisms in cheese (Tsanasidou et al., 2021). The findings of the present study align with those of Demirci et al. (2021), who reported that Enterococcus spp. is the dominant species throughout the maturation of traditional goat tulum cheese, comprising over 60 % of the microbiota at each stage of maturation. Lactiplantibacillus spp. was identified as the second dominant species. Enterococcus and Lactiplantibacillus species were reported to be dominant in Izmir Tulum and Mengen cheeses produced in Türkiye (Karabey et al., 2018; Akoğlu et al., 2017).
Fox et al. (2017) observed a significant occurrence of enterococci in conventional cheeses originating from the Mediterranean region. Enterococcus species are frequently present in the microbial flora of various kinds of cheese made with diverse raw materials and production procedures (Tsigkrimani et al., 2022; Aktaş and Erdoğan, 2022; Albayrak and Duran, 2021; Uymaz et al., 2019; Russo et al., 2018; Milani et al., 2016).
The predominant Enterococcus species found in cheese are E. faecium, E. faecalis, and E. durans. E. faecium and E. faecalis strains, along with other LAB strains, are employed as starter cultures in the dairy sector. The references cited are Graham et al. (2020) and Hanchi et al. (2018). On the other hand, Enterococcus strains pose a major risk in the dairy industry due to their predominance in the human and animal gastrointestinal tract. In addition, the high virulence of some enterococci from dairy products and their resistance to various antibiotics are of great concern. E. faecium and E. feacalis species in particular are thought to be opportunistic pathogens causing various infections. Therefore, it is crucial to correctly identify the species and strains of enterococci found in dairy products (Terzić-Vidojević et l., 2021).
The dominant Enterococcus species identified in our study was E. faecium (86 %). Similarly, a study on Enterococcus species isolated from Ezine cheese reported that they were predominantly composed of E. faecium (64.3 %) (Uymaz et al., 2019).
In the study, Lactiplantibacillus spp. (specifically Lb. plantarum and Lb. pentosus), were found to be the second most prevalent microorganisms in Muş Tulum cheese samples at both early and late stages of maturation. The bacterial diversity in Tulum cheeses was determined using the 16S rRNA sequencing technique, which revealed that the dominant species were Streptococcus and Lactiplantibacillus spp. (Gezginç et al., 2022). The dominant LAB identified in Kargı Tulum cheese produced in Türkiye was Lactobacillus spp. (Lb. paracasei and Lb. plantarum), followed by Streptococcus thermophilus and E. durans (Kunduhoğlu et al., 2012).
Lactiplantibacillus species are industrially significant due to their ability to thrive in low pH and high salt concentrations in cheese, which impacts the cheese’s flavour development (Uymaz et al., 2019). Lb. plantarum has been employed as an additional starter culture in certain cheeses to expedite the maturation (Spus et al., 2017). Our analysis revealed that 90 % of the detected strains of the Lactiplantibacillus genus were classified as Lb. plantarum. Previous studies have reported Lb. plantarum to be the dominant microflora in certain cheeses (Nalepa and Markiewicz, 2022; Özkan et al., 2021; Hassanzadazar et al., 2017).
Lactococcus spp. contribute to curd formation by rapidly acidifying milk and preventing the growth of unwanted microbiota. They also play a crucial role in producing various taste-aroma compounds that contribute to the distinctive flavour of cheese through their strong caseinase, aminopeptidase, and esterase-lipase activities (Gezginç et al., 2022; Pisano et al., 2019). Furthermore, through the metabolism of citrate, they generate aromatic compounds, including diacetyl, acetoin, and acetaldehyde. These compounds are responsible for the distinctive taste of cheeses such as Camembert, Cheddar, and Emmental (Fusieger et al., 2020).
The study identified four isolated Lactococcus strains, comprising of two strains of L. lactis subsp. lactis (FFH51 and FFH54), one strain of L. lactis (FFH48), and another as L. lactis subsp. lactis biovar diacetylactis (FFH53), which accounted for 6.15 % of the total strains. Uymaz et al. (2019) reported that L. lactis accounted for 6.84 % of all strains isolated from Ezine cheese. Muruzović et al. (2018) reported that Lactococcus spp. isolated from traditional Sokobanja cheese consisting of L. lactis subsp. lactis and L. lactis subsp. lactis biovar diacetylactis. Lactococcus spp. is known to be the dominant LAB flora in raw milk and its products, emerging in the early hours of fermentation (Terzić-Vidojević et al., 2020; McSweeney and Sousa, 2000). In Muş Tulum cheese, Lactococcus isolates were detected only on day 45 of maturation and were considered the second dominant species at this stage. According to a study on Tulum cheese, Lactococcus isolates are initially present at low levels at the beginning of maturation (Demirci et al., 2021). Among LAB species, L. lactis subsp. lactis biovar diacetylactis is considered the best flavor producer (Farahani et al., 2017). Therefore, L. lactis subp. lactis and L. lactis subp. lactis diacetylactis species are commonly used as starter cultures to produce commercial cheeses in which milk has been pasteurized.
Only one strain of Pediococcus pentosaceus (FFH20) was isolated from the Muş Tulum cheese samples. While Pediococci do play a role in cheese flavour formation, it is known that their proteolytic and lipolytic roles are not as effective as those of Lactococcus, Lactiplantibacillus, and Enterococcus species (Uymaz et al., 2019). In Feta and Teleme cheeses, P. pentosaceus isolates have been reported to exhibit slow acid formation and produce more diacetyl and acetaldehyde (Litopoulou Tzanetaki and Tzanetakis, 2011). Several studies have reported low levels of P. pentosaceus in certain types of cheese (Tsigkrimani et al., 2022; Gantzias et al., 2020; Shi et al., 2019; Şenocak Soran, 2018; Ertürkmen and Öner, 2015).
Technological characterization of LAB strain
Acidification capacity of strains
Table 4 displays the pH and titratable acidity alterations of the 65 isolated strains at the 3rd, 6th, 9th, and 24th hour. LAB strains can be categorized according to their acidification ability, which is measured by the increase in acidity of skim milk over a period of 24 hours (Anagnostopoulos et al., 2018). Based on this classification, it was found that all Lactococcus strains (FFH48, FFH51, FFH53, and FFH54), as well as E. faecium FFH2 and Lb. plantarum FFH59 strains demonstrated a high acidification capacity (reducing pH by more than 2 units). The remaining 29 E. faecium, 4 E. faecalis, 2 E. durans, 1 P. pentosaceus, and 9 Lantiplantibacillus strains exhibited a moderate acidification capacity (reducing pH by 1.5-2 units). Additionally, 13 E. faecium and 1 E. durans strain showed a low acidification capacity (reducing pH by less than 1.5 units). LAB isolates with a high acidification effect are important for developing the desired taste and aroma in produced cheeses and preventing spoilage-causing bacteria growth. According to Aspri et al. (2017), bacteria that cause fast acidification should reduce the pH of milk to a level below 5.3 within 6 hours at a temperature of 37 °C. The study found that only L. lactis strains (FFH48, FFH51, and FFH53) were rapid acid producers, reducing the pH below 5.3. In contrast, the other strains exhibited moderate or slow acid production. It is well-known that lactococci have a higher acidification capacity, particularly in the first 6 hours of incubation, than lactobacilli and enterococci, due to their rapid metabolization of lactose (Pisano et al., 2019; Turhan and Öner, 2014).
Table 4. Acidification capacity (pH and % L.A.) and proteolytic activity (P.A.) of LAB isolated from Muş Tulum cheese
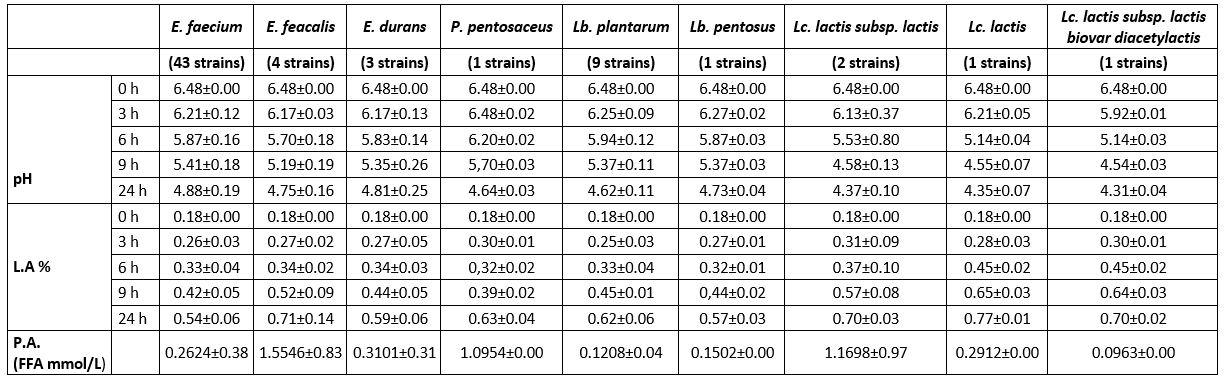
pH, L.A., and P.A. the values are the average values of the strains.
Previous research has reported that isolated L. lactis species exhibit a rapid acidification capacity (Akoğlu et al., 2017; Kırmacı et al., 2016; Ertürkmen and Öner, 2015), while enterococci demonstrated moderate and low levels of acidification capacity, consistent with some studies (Anagnostopoulos et al., 2018; Ribeiro et al., 2014). All Lb. strains, except one, demonstrated a moderate level of acidification capacity, similar to our study. Elçioğlu (2010) suggested using Lb. plantarum, E. faecium, and E. durans strains with rapid acidification capacity as starter cultures for Kargı Tulum cheese. An Lb. plantarum strain isolated from Algerian cheese was reported to have high acidification capacity (Metrouh et al., 2022).
In addition, according to Herreros et al. (2003), strains that produce at least 0.25 g of lactic acid per 100 mL of milk after 6 hours of incubation are considered appropriate as starter cultures for cheese manufacturing. Following incubation, titrimetric method (Sarantinopoulos et al., 2001a), the L. lactis strains (FFH48, FFH51, and FFH53) and the E. faecium FFH50 strain generated a lactic acid produced of 27 mg/mL. Based on these findings, strains that exhibit a high capacity for acidification and rapid acid generation are deemed appropriate for utilization as primary or adjunct starter cultures in cheese manufacturing.
Proteolytic activity levels of strains
LAB demonstrate effective proteolytic activity through their proteinase and peptidase activities. LAB has a substantial impact on the creation of different aromatic compounds that affect the taste of cheese. This is achieved by transforming casein into smaller peptides and amino acids (Nicosia et al., 2023). Excess proteolysis in cheese can lead to unpleasant flavours. This is caused by the interaction of peptides and free amino acids (FAAs) with other chemicals (Duan et al., 2019).
The proteolytic activity of the strains was analysed using the OPA technique, and the results showed that the activity ranged from 0.0347 to 2.2452 mmol/L of FAA (Table 4). Out of the 65 strains tested, 57 exhibited low levels of proteolytic activity, with FAA levels less than 1 mmol/L. Strains E. faecium (FFH13 and FFH45) (1.1091 and 1.2147 mmol/L), E. faecalis FFH69 (1.5996 mmol/L), and P. pentosaceus FFH20 (1.0954 mmol/L) showed moderate levels of proteolytic activity (FAA between 1-2 mmol/L). The highest proteolytic activity (FAA more than 2 mmol/L) was observed in E. faecalis (FFH46 and FFH14) (2.2452 and 2.1926 mmol/L), L. lactis subsp. lactis FFH51 (2.1484 mmol/L), and E. faecium FFH16 (2.0593 mmol/L) strains, respectively. In contrast to other enterococcal strains, E. faecalis strains have been found to have high levels of proteolytic activity. This finding aligns with other research documenting elevated proteolytic activity (>2 mmol Leu) in E. faecalis strains relative to other enterococcal strains (Sarantinopoulos et al., 2001b). However, the Lb. plantarum strains demonstrated minimal proteolytic activity ranging from 0.0834 to 0.2088 mmol/L, consistent with the findings of Belarbi et al. (2022) (0.0-0.1460 mg Leu/ml). Akoğlu et al. (2017) show that lactococci have higher proteolytic activity than enterococci and lactobacilli. The study discovered that only one Lactococcus isolate (strain FFH51) exhibited high proteolytic activity.
Bacteriocin activity of strains
Enterococci exhibit antimicrobial activity against pathogenic microorganisms that cause food spoilage thanks to the bacteriocins (enterocins) they produce. This allows some enterococci strains to be used as starter cultures in cheese production. The results showed that only two Enterococcus strains (E. faecium FFH12 and E. faecium FFH77) exhibited inhibition activity against Lactococcus lactis SIK1403, indicating that these strains are bacteriocin producers (refer to Figure 3). E. faecium and E. durans strains isolated from raw milk are reportedly used as adjunct cultures in producing Izmir Tulum cheese (Yerlikaya and Akbulut, 2019). Therefore, tests regarding factors such as antibiotic resistance, virulence factors, and hemolytic reaction must be performed before Enterococcus strains isolated in the production of Muş Tulum cheese are approved for use as starter cultures.
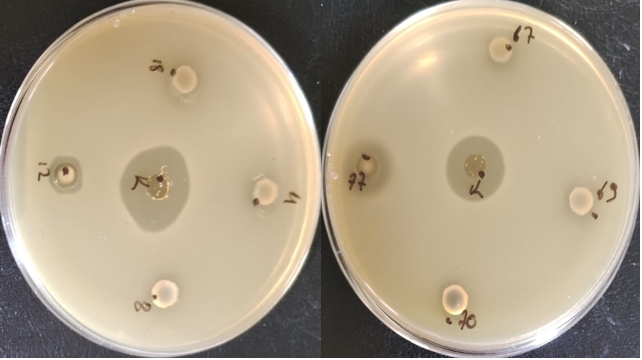
Figure 3. Antimicrobial activity of E. faecium species against L. lactis. K: P. pentosaceus OZF strain, 4: Enterococcus faecium FFH4, 8: Enterococcus faecium FFH8 12: E. faecium FFH12 strain, 18: Enterococcus faecium FFH18, 67: Enterococcus faecium FFH67, 69: Enterococcus faecalis FFH69, 70: Enterococcus faecium FFH70, 77: E. faecium FFH77 strain
Conclusion
Indigenous LAB present in raw milk play a significant role in determining the sensory characteristics of cheese. Identifying and incorporating these bacteria into the cheese industry is of great significance. This study aims to identify the dominant lactic bacteria strains present in Muş Tulum cheeses produced from unpasteurized sheep's milk throughout the storage period using biochemical, phenotypic, and genotypic methods. The LAB distribution isolated from Muş Tulum cheese was classified as Enterococcus spp. (76.92 %), Lactiplantibacillus spp. (15.38 %), Lactococcus spp. (6.15 %), and Pediococcus spp. (1.54 %). Based on technological characterization, strains that exhibit high acidification or proteolytic activity, such as L. lactis (FFH48, FFH51, FFH53, and FFH54), Lb. plantarum FFH59, E. faecium (FFH2 and FFH16), and E. faecalis (FFH14 and FFH46), are considered to have the potential to be used as starter or mixed cultures in Muş Tulum cheese production. As a result of antimicrobial activity, which is one of the essential criteria in the selection of stater culture, it was determined that 2 E. faecium strains (FFH12 and FFH77) were bacteriocin producers. In addition, the isolated L. lactis strains could potentially be used in Muş Tulum cheese production, and their impact on the cheese's technological properties may be investigated in a subsequent study.
Funding
This study was funded by Harran University Scientific Research Projects Authority (HUBAK Project No.: 23067).
References
https://tez.yok.gov.tr/UlusalTezMerkezi/ tezSorguSonucYeni.jsp.
Sarantinopoulos, P., Kalantzopoulos, G., Tskalidou, E. (2001): Citrate metabolism by Enterrococcus feacalis FAIR-E229. Applied and Environmental Microbiology 67 (12), 5482-5487.
