Introduction
Amyloidogenic proteins are well known for their ability to misfold and aggregate normal proteins into insoluble fibrils. It plays a crucial role in the pathogenesis of several neurodegenerative diseases, including Alzheimer's, Parkinson's, Huntington's, and other diseases like diabetes type II. This aggregation of amyloid fibrils disrupts cell function, resulting in gradual tissue impairment. To degrade these aggregates, the body employs a range of proteolytic enzymes. Serine proteases have created widespread interest due to their broad substrate specificity and potent catalytic activity [1]. On the other hand, evidence exists that amyloids can be degraded in vitro using zinc oxide nanoflowers [2] and nanoformulated carrageenan [3].
Hydrolytic enzymes, such as proteases, phytases, lipases, amylases, amidases, and others, dominate the enzyme market today. Most of the proteases belong to the serine protease family and are characterized by a serine residue [4]. Serine protease acts as an endopeptidase by breaking down peptide bonds like other proteins. However, these residues in active sites play a crucial role in coordinating functions through protein hydrolysis [5]. Moreover, these types of protease are widely distributed in every living organism, including viruses, bacteria, fungi, insects, and humans, and function both within and outside the cells [6]. Serine proteases are traditionally classified into various categories, including chymotrypsin-like, trypsin-like, subtilisin-like, elastase-like, and thrombin-like protease. Chymotrypsin-like proteases can be found in both prokaryotes and eukaryotes. Over 240 chymotrypsin-like proteases have been identified in the MEROPS peptide database. Chymas, synthesized by mast cells, play essential roles in various peptide hormones and other physiological processes, such as hemostasis, inflammation, apoptosis, signal transduction, immunological responses, and digestion. It also breaks down the peptide bonds on the carboxyl side of tryptophan, phenylalanine, and tyrosine. This particular type of protease sequentially activates blood clotting, fibrinolysis and wound healing [7]. Trypsin-like protease is a homologous protease found in the human genome. Out of 699 proteases in humans, 138 belong to the S1 protease family and the other 178 are serine proteases [8]. This trypsin-like protease performs a key function in boosting the body’s response to blood clots, protein digestion, and immunity [9]. Subtilisin is a serine protease found in prokaryotes. According to the MEROPS database, subtilases are a subfamily of the clan SB of serine peptidases and belong to S8A family. Generally, they act as a detergent to remove proteinaceous dyes. Compared to other protease types like trypsin and chymotrypsin, elastase-like protease has a smaller S1 cleft. These proteases cleave bonds at the non-polar amino acids such as alanine, glycine, and valine [5,10]. Snake venoms contain numerous highly toxic thrombin-like serine protease isoforms. It consists of two chains linked by disulfide bonds: the A and B chains. It is involved in fibrinolysis, and Alzheimer's and Parkinson's-induced dementia and other neurodegenerative conditions [5,11].
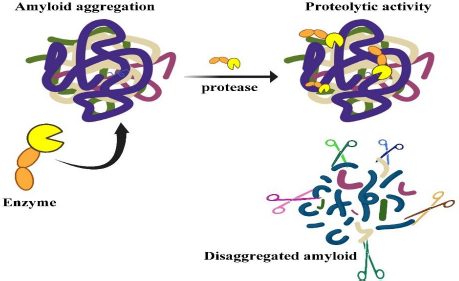
Among those serine proteases, lumbrokinase (LK) and serratiopeptidase (SP) are the world's most exciting enzymes being studied for their potential application in the medical field [5]. A new dimension has emerged with the discovery of nattokinase (NK) and keratinase (KER), which are unique proteases that are capable of destroying recalcitrant proteins like amyloid, known as protease-resistant proteins [12,13]. The mechanism of protease in the clearance of amyloid misfolded proteins is depicted inFigure 1.
Specific types of serine proteases
Lumbrokinase
Since ancient times (<4000 years, 2600 B.C.), earthworms have been closely linked with human beings. They have been utilized as food and medicine to treat various human ailments in different parts of the world, including China, Japan, Korea, India, Cambodia, Myanmar (Burma), Vietnam, Iran, and the Middle East [14]. The Greek philosopher Aristotle (384-322 B.C.) mentioned the importance of earthworms and termed them “The Intestine of Earth” [15-17]. They also have been used for sore throat, healing wounds, piles, chronic boils, and so forth for an exterior application, and traditional treatment of internal diseases like chronic cough, diphtheria, jaundice, rheumatic pains, tuberculosis, bronchitis, facial paralysis, and impotence [18].
The epithelial cells in the gizzard segment, especially in the anterior alimentary regions, express and produce earthworm fibrinolytic proteases. In these parts, the proteases possibly play a role in digesting protein and peptides in food [19]. LK was isolated chiefly from Lumbricus rubellus, and Eisenia fetida. Earthworm protease enzymes (EPE) and fibrinolytic enzymes (EFE) are some names given to LK [20]. In some cases, proteases are named after the Latin binomial for the species of earthworm from which they are derived. For example, E. fetida protease (Efp) is a protease extracted from E. fetida. Several fibrinolytic enzymes were extracted from L. rubellus, including F-1-0, F-1-1, F-I-2, F-II, F-III-1, and F-III-2 [21]. Zhou et al. purified seven fibrinolytic proteins from E. fetida in 1988 [22]. LK isozymes were also extracted from L. biomass and E. Andrei. LK can sustain its activity in both basic and acidic conditions, having a wide pH range from 1 to 11. The molecular weight of LK lies between 20 and 35 kDa, with a range of isoelectric pH (pI) from 3 to 5 [21]. Moreover, some LK are resistant to high temperatures and can withstand up to 60° C. The same fibrinolytic enzyme has multiple names because each enzyme was isolated and named independently by different research groups, making accurate determination of the total number of LK difficult. Nomenclature for LKs should be standardized according to their function, properties, and sources [23]. Conventional approaches to LK extraction and earthworm purification are complex and time-consuming. In order to assess the potential clinical application of LK protein, researchers used recombinant DNA technology to express and characterize a single LK protein [24]. The plasminogen activation system mechanism of LK differs from other thrombolytic enzymes, such as streptokinase, urokinase, staphylokinase, recombinant tissue-type plasminogen activator, and recombinant prourokinase. LK is not only involved in the activation of plasminogen but can also activate fibrin directly [25]. These enzymes execute predominantly proteolysis of fibrinogen and fibrin and hardly hydrolyse other plasma proteins, such as plasminogen and albumin [25,26].
According to the data available in GenBank, eight clones of accessible cDNA of LK (GenBank Accession No.; AY187629, U25644, AY438622, AY178854, AF304199, U25648, AF433650) are available. The coding sequence of LKs cDNA has a length of 852 bp and encodes 283 amino acids, in which 36 amino acid codes for the signal peptide and the remaining 247 amino acids code for the whole protein [24,27,28]. The nucleotide arrangement in each clone of cDNA starts with 13 codons containing "CG" motifs in the complete sequence, which is almost rare for mammals. This makes the translation of such DNA in mammalian cells or tissues inefficient [29]. A cDNA of gene EFE-3 of Eisenia fetida has 859 independent nucleotides, with an open reading frame accessible from 112-853, which encodes a polypeptide with 247 amino acid residues [27].
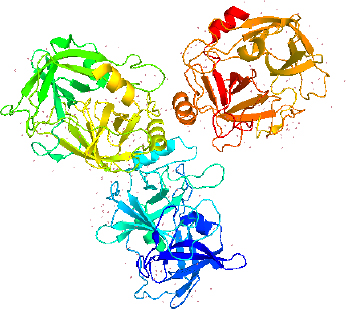
The sequence of LK isolated from different earthworm species was analysed to get the full proteomics data. The protein sequence from L. rubellus and E. fetida had more residues than the secondary structures, including α-helix, turn, β-sheet, and coil. The arrangement of an LK isoform known as earthworm fibrinolytic protease II (Ef P-II (EFEa)) is remarkably identical with other serine proteases with its known structures [30,31] or serine protease related to earthworm [29]. The earthworm protease catalytic characterization is directly affected by its tertiary structure. The studies on NMR and X-ray analysis revealed that Ef P-III-1 (EFE-b) belongs to the group of trypsin-like protease possessing two chains- a light chain at the N-terminal of pyroglutamate and an N-glycosylated heavy chain [32]. The structural aspect showed Ef P-III-1 having high stability regarding heat resistance and resistance to proteases and organic solvents [33]. Further, it was noted that another LK isoform, Ef P-II is a serine protease type chymotrypsin possessing an essential S1 binding pocket [31].
LK structure extracted from PyMol with PDB id 1M9U, as depicted inFigure 2, showed the chymotrypsin-like serine proteases with characteristics of S1 elastase. The S1 specificity of the beta strand significantly showed four different residues (Ser-Ser-Gly-Leu) after Val217, which can provide additional substrate for the hydrogen binding sites for distal P residues and gives elongation for the S1 pocket. The S1 pocket prefers elastase-specific small hydrophobic, strong, and bulky P1 residues that support the tight substrate binding and induced S1 pocket fitting. This structure was first labelled to represent earthworm fibrinolytic enzyme components as well as serine protease, which originated from the annelid worms [34].
Fibrinolytic and proteolytic activities of lumbrokinase
Lumbrokinase (LK) has been utilized as a fibrinolytic agent in China, Japan, and Korea. The arrangement of thrombus inside the vessels of blood causes numerous inconveniences, for example, myocardial infarction and stroke. A considerable lot of local and recombinant proteins with proteolytic properties have been utilized as effective thrombolytic agents, such as urokinase (UK), streptokinase, recombinant tissue-type plasminogen activator, recombinant prourokinase, and staphylokinase [35-38]. A considerable lot of them have indicated great outcomes. However, they additionally have a few confinements, for example, quick clearance, the absence of protection from reocclusion, bleeding complications, and other adverse impacts [39]. LK has both functions, like plasminogen activation and fibrinolysis and has been used to treat thrombosis [40]. When consumed orally, the animal experiments showed that LK had a significant fibrinolytic efficacy. Additionally, distinct improvement was witnessed in the management of high blood viscosity syndrome and thrombocytosis [41]. Moreover, LK is relatively stable under long-term storage at ambient temperature. As an oral medication, LK is known to avert and treat clotting diseases, including cerebral thrombosis and myocardial infarction [42]. Specifically, Lumbricus rubellus and Lumbricus bimastus, are the essential resources for a fibrinolytic agent in Southeast-Asian nations, for example, China, Korea, Japan and so on [43,44]. The isolation and purification of EFEs included centrifugation, filtration, ammonium sulphate precipitation and dialysis, ion exchange chromatography, and affinity chromatography [45], which made the scaling up difficult. A simple method was employed to scale up, like aqueous-two phase systems (ATPS), which included separation and the target protein concentration, giving a clear extract [46]. Seven types of purified fibrinolytic enzymes (EFE-a to EFE-g) and eight glycosylated purified fibrinolytic enzymes (EfP-0-1 to EfP-III-2) were identified from the source of Eisenia fetida till today [21,47,48].
Six fractions of lumbrokinase (F-I-0 to F-III-2) having fibrinolytic activity are purified from Lumbricus rubellus lysates using autolysis, fractionation with ammonium sulphate and column chromatography. Lumbrokinase’s F-III-1 and F-III-2, F-I-1and F-I-1 both showed a similar activity although they have different molecular weights [23].Table 1 summarizes the various physiological and biological properties of these enzymes.
There has been an increase in the use of the LK protein in the field of medicine today, mainly due to its ability to perform thrombolytic therapy. Because LK is a eukaryotic protein, it is even more important in the treatment of cardiovascular and cerebrovascular diseases. LK has been produced both in bacterial and eukaryotic systems as a result of advances in recombinant technology. As a result of advanced design of the appropriate host system and alteration of the genetic level of a drug to make it more effective, there have been attempts to improve the expression of the recombinant enzyme, immobilization for reuse of the enzyme, and chemical modification in order to reduce antigenicity and improve its activity in order to make a drug more effective. It is expected that it will be the right drug candidate in the future for the treatment of thrombosis [49]. Moreover, an in vivo study demonstrated that the fibrinolytic enzyme from earthworm has reduced intra-abdominal adhesion and peritoneal thickening [50].
Role of lumbrokinase in cerebral Ischemia and Implantation
By increasing the level of c-AMP and reducing calcium release from calcium reserves, LK has anti-ischemic activity due to its antiplatelet activity. LK had an antithrombotic effect due to the inhibition of expression of ICAM-1 and an antiapoptotic effect due to the activation of the JAK1 / STAT1 pathway, which causes the antithrombotic effect. As a result of the antithrombotic and antiapoptotic functions of the Janus Kinase 1 / signal transducers and transcriptional activators 1 (JAK1 / STAT1), the brain was protected from ischemic damage through the action of ICAM-1 and Janus Kinase 1 on the brain's intercellular adhesion mechanisms [51]. When an artificial organ is transplanted, a small thrombus usually forms on the surface of the graft, which may lead to serious complications, such as the rejection of the graft if the thrombus is not removed. Despite the fact that there has been considerable achievement in the field of medicine, there is still a concern about the compatibility of the organ with the improvement in blood circulation, which leads to offensive results in transplantations. Maleic anhydride-methyl vinyl ether (MA-MVE) has been used as a catalyst for immobilizing the LK on polyurethane and, as a result, has shown high anti-thrombogenic activity, and has reduced the formation of surface-induced thrombi. There is a possibility that an immobilized LK surface may reduce the adhesion and activation of platelets by inhibiting fibrinogen adsorption or by altering the conformation of adsorbed fibrinogen at an early stage of blood contact by decreasing their adhesion and activation [52]. The oral administration of LK led to a significant increase in the levels of silent information regulators (Sirt1), respectively. The study report suggested that the post-ischemic treatment with LK from earthworm mitigated myocardial IR injury by stimulating Sirt1 and, enhancing autophagic flux, and reducing IR-induced injury [53]. Another study conducted by Wang et al. evaluated that LK (10 μg/kg) exhibits protective effects against heart disorder in a rat model [54]. This collection of evidence could successfully conclude that LK has promising applications in biomedicine.
Serratiopeptidase
Serratiopeptidase (SP) (EC number 3.4.24.40) is an active proteolytic enzyme found in enterobacteria Serratia marcescens (Gram-negative bacteria) and belongs to the family of alkaline serine protease. It has a molecular weight of 45 kDa to 60 kDa and comprises three zinc atoms as ligands at the active site [55]. SP cannot bind proteins in healthy tissues since its chemical structure prevents it from doing so [56]. SP depends on secretory protein on the membrane for its production, and the N-terminal signal peptide-independent pathway secretes it [57]. SP is considered by high-level specificity towards its substrate. The fibrinolytic enzyme activity of SP is due to its activity in the degradation of insoluble proteins, such as fibrin and other mediators of inflammation [58]. They were also used to treat several chronic disorders like arthritis, atherosclerosis, bronchitis, fibrocystic breast disease, carpal tunnel syndrome, Crohn’s disease, traumatic swelling, leg ulcers, fibromyalgia, migraine, breast engorgement, sinusitis, lung disorders, hepatitis, diabetes, thrombosis, carotid artery blockage, and uterine fibroids [59,60]. This enzyme is available as a supplement to enhance the cardiovascular system and overall health. The enzyme SP is also considered to be a healing enzyme because it heals sprained muscles, traumatic injuries, leg ulcers, and torn ligaments, drains mucus, postoperative inflammation, and reduces the viscosity and elasticity of nasal mucus [61].
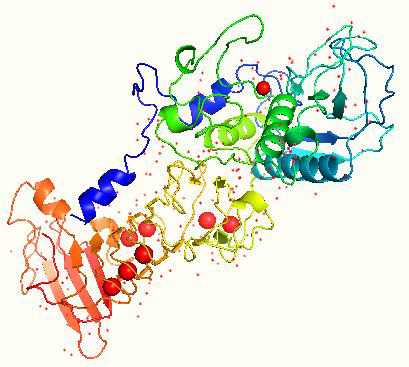
The gene encoding SP is composed of 470 amino acids. SP amino acid sequence is free from sulfur, methionine, and cysteine [62] with a G + C content of 58 % [63]. The maximum SP activity is at pH 9.0 and a temperature of up to 40 °C [58]. The saturation point where SP is thoroughly degraded was found to be at 55 °C [62]. The isoelectric point of SP is found to be 5.3 [64]. The gene sequence of SP is cloned and sequenced, and its crystal structure has been determined, as shown inFigure 3 [63].
Serratia marcescens HR-3, SP gene was expressed in E. coli (DE3) / pLysS using the pET32a (+) expression vector. It has been reported that the enzyme is highly expressed as inclusion bodies and that purified SP contains no activity [65]. Similarly, the SP gene from Serratia marcescens strain SM6 was allowed to express in E. coli by using the lac promoter, which secreted the enzyme into the medium, but the enzyme was found to be inactive with a high molecular weight [66]. The structural gene of SP is not clustered with the secretion genes, which makes improper processing of the zymogen protease that is dysfunctional in the E.coli expression system, making the recombinant SP inactive [67]. Further, the SP gene was cloned into the pPICZαA Pichia expression vector and electrotransformed into the Pichia pastoris GS115, outputting the maximum expression at 72 h using the E.coli expression system. Yeast can be a suitable alternative host for such types of secretory proteins [31].
Fibrinolytic activity
SP is known to be safe as it is regularly taken by the oral route as a fibrinolytic drug and has the potential to circulate in the blood due to its ability to be absorbed by the intestine [68,69]. The therapeutic use of SP may lead to less bioavailability because of its lower membrane permeability and high tendency to undergo degradation by the enzymes present in the gastrointestinal tract [69]. In rats, after taking a dose of 100 mg/kg of SP, SP can be detected in the serum and lymph at concentrations of 0.87 and 43 ng/mL, respectively [32,33]. The exact bioavailability data of SP in humans is not yet reported anywhere in the literature [69].
Fibrin can repair injuries caused by trauma, surgery and damage to old cells, replacement by new cells, tissues, and muscles [70]. The blood vessels of the damaged tissue form a composite substance called thromboplastin, which binds to the broken blood vessels of the tissue and emits a platelet factor if the blood vessels are damaged. A Prothrombin activator is created when thromboplastin and platelet factor react with calcium ions and other elements to form an insoluble fibrin that blocks blood flow and oxygen supply in the body, causing infarction of myocardium, stroke, pulmonary embolism, and vein clots [59]. Thus, fibrinolytic enzymes like SP, which dissolve clots, are crucial to the human body to avoid the considerable risk of damage. Mei et al. [71] demonstrated strong thrombolytic effects in in vitro model, with 96.6 % clot lysis at a concentration of 300 U/mg of SP. Therefore, the study indicates that SP could be a suitable candidate for both the prevention and therapy of thrombotic disorders [71].
Anti-inflammatory effect of serratiopeptidase
Most diseased conditions produce a certain level of inflammation, which causes hostility in the disease. This inflammation aggravates the immune system to activate white blood cells (WBCs), which travel via the circulatory system to destroy pathogenic bacteria and foreign substances and kills cancer cells. During the repair mechanism, the WBCs can translocate into organs or tissues, and such elevated activity of WBCs results in cell damage [61]. SP was initially utilized for its anti-inflammatory properties in Japan in 1957 [56]. Khateeb et al. [72] examined the anti-inflammatory impact of SP on orthodontal inflammatory syndrome. In general, pathogen-induced inflammation can manifest as either acute or chronic inflammation. Acute inflammation is a protective response to infection, while uncontrolled resolution leads to chronic inflammation [72]. The remarkable anti-inflammatory effects of SP and diclofenac in acute and subacute inflammation were evaluated by Jadav et al. [73]. Nonsteroidal anti-inflammatory drugs (NSAIDs) are recommended for acute inflammation, while NSAIDs, in combination with steroidal drugs and SP, are used to address chronic inflammation [74]. Recent investigations established that SP combined with paracetamol can successfully reduce edema and acute inflammation in rat models. Hence, this study also suggests that SP could be a promising candidate for anti-inflammatory drugs [75]. A study by Krishna et al. also highlighted the anti-inflammatory activity of SP, demonstrating its effectiveness in reducing postoperative swelling compared to dexamethasone [76]. The role of SP in vascular inflammation induced by lipopolysaccharide (LPS) was also investigated. The result showed that SP reduced LPS-induced oxidative stress in mice and decreased the expression and activity of monocyte chemoattractant protein-1 (MCP-1) [77]. The author evaluated the effectiveness of two drug combinations for reducing postoperative pain and swelling after the surgical removal of impacted mandibular third molars. In this study, two drug combinations were orally administered to sixty patients three times continuously for three days. The drug combination of trypsin, rutoside, and bromelain showed less swelling. Whereas diclofenac and serratiopeptidase drug exhibited higher swelling and less pain. Hence, the study concluded that bromelain drug is effective in reducing swelling, and diclofenac, SP-based therapy provides superior pain relief [78]. The anti-inflammatory effect of SP ointment was evaluated in mice. Compared to control group, SP ointment treated mice demonstrated significant anti- inflammatory effect [79].
Clinical applications of serratiopeptidase
SP reduces pain by arresting the discharge of pain, which influences amines such as bradykinin from inflamed tissues [80]. SP has the capacity to cause hydrolysis of histamine, serotonin, and bradykinin, which are accountable for eliciting oedemic responses [81]. The substrate bradykinin binds in the vicinity of the zinc-binding site of SP, which efficiently cleaves the peptide bond of bradykinin [82]. At the infection site, they modulate the transformation of cell adhesion molecules, which are intricate in the inflammatory regulatory cells. SP is taken orally to control the inflammation that occurs due to sinusitis, prostate gland inflammation, carpal tunnel syndrome, breast engorgement, chronic emphysema, and acute and chronic ear-nose-throat disease [55]. In 2021, Gupta and colleagues illustrated the efficacy of using SP and vitamin D together to combat the severe consequences of COVID-19 conditions [83]. During the pandemic, COVID-19 has led to various respiratory diseases, including nasal congestion and cough among patients. Various studies reported that mucolytic drugs can help boost bronchial mucus secretion or reduce mucus viscosity [55]. Furthermore, a comparison of SP’s mucolytic activity with seaprose indicated that proteolytic enzyme were active in in vivo animal models [84]. Gioia’s 2020 study on patients with respiratory disease found that SP enhanced mucus clearance by lowering the neutrophils, which changed sputum viscosity. This shows that SP could be an effective therapy for respiratory problems and COVID-19 consequences [85].
The bacterium thrives by making biofilm within the microbiome. These biofilms usually form surfaces of the bacteria; for example, Staphylococcus aureus, which has a variety of virulence factors and possesses the potential to invade the eukaryotic cells by making surface biofilm and dominates with staphylococcal infections. Blocking of such Staphylococcus aureus colonization can inhibit the quick spread of biofilm [86].
The anti-infective effect of SP is exerted through inert surface attachment, thereby inhibiting Staphylococcus adherence and ceasing invasion of eukaryotic cells. The detailed mechanism is yet to be explored. SP has also shown an antibacterial effect against Escherichia coli and Pseudomonas aeruginosa [87,88]. Mecikoglu et al. [89] conducted an in vivo study that used SP to treat bacterial infections. 94.4% of the infected rats treated with SP recovered, showing its potential in combating biofilm-forming activity. Using proteolytic enzymes like SP in combination with antibiotics could be effective against microbes [89].
Keratinase
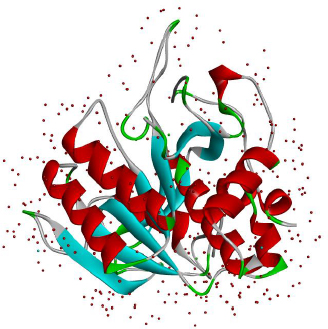
Keratinases (KER) are extracellular enzymes usually produced from Bacillus, Streptomyces, Aspergillus, Fervido bacterium, Xanthomonas, Chryseo bacterium, and Vibrio, which are known to degrade keratin protein in eukaryotes. KER is especially known for its use in de-hairing leather before processing and in the cosmetic industry [90,91]. Prion disease or transmissible spongiform encephalopathy (TSE) is a neurological disease manifested due to the aggregation of misfolded proteins called prions. Prion protein gets transmitted from infected animals through the consumption of cattle feed prepared from dried brains and carcasses of dead infected animals [92,93]. The types of TSEs in humans are CJD, transmissible mink encephalopathy (TMC) in mink and kuru [94]. A reported novel application of KER (also known as prionzyme) is the hydrolysis of the pathogenic form of prion protein, which is known to be resistant to any other protease [95]. The present prion treatment methods comprise its inactivation using chemicals and heat, which has limitations regarding environmental acceptability, application and cost compatibility, making way for enzyme inactivation of prion [96]. KER-mediated prion protein destruction is feasible at a nominal cost and is eco-friendly [97]. The structure of keratinase is extracted from the Protein database and given inFigure 4.
Amyloidogenic activity of keratinase
Amyloid β peptides (Aβ) are poisonous peptides composed of 39 to 43 amino acid residues. When Aβ aggregates are deposited in brain tissues, they block synaptic transmission and cause neurodegenerative illnesses. Several experiments have been done to disintegrate these hazardous aggregates. Amycolatopsis Ker1 and Ker2 were tested for their ability to break down Aβ fibrils. In this study, Aβ fibrils of lysozymes were treated with varying doses (50, 100, 125, 250, 500 mg/ml) of keratinase, and their degraded structure was analysed over time intervals. At 24 h, Ker1 had a larger effect than Ker2 [99]. In recent years, numerous studies have suggested that keratinolytic enzymes could potentially be utilized in degrading Aβ aggregates and prion protein (PrPSc). Keratinase MSK103 from Bacillus licheniformis efficiently degraded PrPSc in brain tissue homogenates infected with scrapie in 2 h at 50 °C. After 20 hours of decontamination, prions were reduced to lower levels. Hence, it proves that MSK103 keratinase can also degrade PrPSc [100]. The researchers discovered that the combination of B. licheniformis N22 keratinase and Pseudomonas aeruginosa NCIMB 8626 biosurfactant formulation was capable of partially degrading ME7 scrapie prions after treating them for 1 h at 50 °C. When the temperature increased from 50 to 65 °C, the PrPSc proteins became undetectable in just 10 min. Interestingly, it was noted that the B. licheniformis N22 keratinase alone could not completely break down the ME7 scrapie prions even after extending the period for 1 h. However, the biosurfactant NCIMB 8626 combined with enzyme N22 keratinase showed complete degradation of infectious prion protein [101]. KER isolated from the moderate thermophile B. licheniformis PWD-1 strain was the first described enzyme that was capable of disintegrating pretreated prion (i.e., heated at 115 °C for 40 min along with SDS) [102]. The purified version of KER (strain MSK 103) showed the degradation and decontamination of prion-infected brain homogenates at 50 °C without prior heat treatments with detergents. This KER has shown intense activity in the 60-70 °C range with pH 9 to 10 [103]. Using an alkaline serine protease isolated from Streptomyces 99-GP-2D-5 strain, scrapie prion degradation could be achieved within 3 min. At a temperature of 60 °C and a pH level of 11, the enzyme has been shown to exhibit its maximum activity [104]. Therefore, most studies concluded that combination therapy of keratinase enzyme has enhanced anti-amyloidogenic activity against Aβ and prion [105]. The limitation of keratinase action is the high temperature it needs to degrade the amyloids in most cases, which cannot be achieved under physiological conditions. Keratinase can be used for surface sterilizations and sterilizations of surgical equipment.
Nattokinase
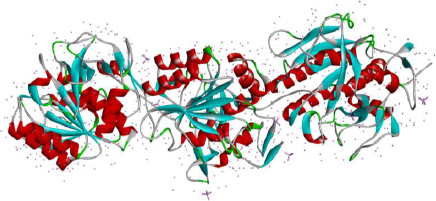
The consumption of boiled soybeans has been practiced for almost 100 years in different Asian regions. The microbe present in fermented natto is a Gram-positive bacteria that can also produce endospores and is named Bacillus subtilis natto (formerly known as Bacillus natto) [106,107]. Nattokinase (NK) is known as Subtilisin NAT, an extracellular enzyme produced by Bacillus subtilis natto [108]. NK comes into the serine protease family with the catalytic triad that is conserved with three residues like Asp-32, His-64, and Ser-221 [109]; it has an isoelectric point at pH 8.7, and the molecular weight is about 27.7 kDa [110]. NK is made up of 275 amino acids, and its gene sequence is identical with various members of the subtilisin family (86 % with subtilisin BPN, and 72 % with Carlsberg subtilisin type, 99.5 % homology with subtilisin E). It disintegrates fibrin in thrombi as well as cleaves plasminogen activator inhibitor type I [111]. The structure of nattokinase from the protein data bank is shown inFigure 5.
NK supersedes plasmin toward thrombolytic activity [112], and oral administration causes absorption from the intestinal tract to induce fibrinolysis. This makes NK a potential anticoagulant for cardiovascular disease treatment and coronary artery disease [113]. The nutritional supplement natto helps to destroy the intima of arteries thickening and aids in the breakdown of mural thrombi seen as a result of endothelial injury [66].
The results showed a reduction in fluorescence emission at 487 nm of Thioflavin-T (ThT) dye (a dye that specifically binds to amyloids), negative ellipticity at 218 nm in Circular dichroism (CD) spectrum, and less fibril morphology in electron microscopic observation. Overall, this study explored the potential of NK for the degradation of amyloid at a temperature of 40 °C and pH 7 [12].
Fibrinolytic of nattokinase
Nattokinase (NK) has been recognized for its fibrinolytic activity by disintegrating amyloid at normal body temperature and neutral pH. This makes it a promising candidate for treating amyloid-related diseases and other neurodegenerative diseases. Numerous studies have reported that both in vitro and in vivo NK has neuroprotective properties [115]. For instance, oral administration of NK has been shown to be effective against cerebral ischemia, resulting in reduced infarct quantity in gerbils due to enhanced fibrinolytic activity [116]. The enzyme NK not only dissolves blood clots but also breaks down Aβ. The study result showed fibrinolytic activity in NK that has lowered factor VII and factor VIII [117]. The study conducted by Guangbo et al. revealed that the NK extracted from B. subtilis has significant fibrinolytic activity. The recombinant NK displayed a clear thrombolytic effect and exhibited strong pH and thermostability [118]. NK has exhibited strong clot-dissolving capabilities and a low risk of bleeding. It reduces clot lysis time and lowers the indicators linked to cardiovascular disease [119]. The NK extracted from B. subtilis G8 has shown excellent fibrinolytic activity [120].
Kamiya et al. found that the NK enzyme reduced the length of carrageenan-induced rat tail and the mass of ferric chloride-induced carotid thrombus [121]. The cationic peptide from natto extract exhibited excellent angiogenic activity in human umbilical vein endothelial cells, indicating the potential of NK as an antithrombotic agent [122]. Another study reported that sustained release of synthesized NK-conjugated magnetite nanoparticles was identified as a potential candidate for thrombolysis [123]. A recent study showed the application of NK towards amyloid degradation by efficiently degrading the three types of in vitro prepared amyloid fibrils-insulin, Aβ1-40, and huPrP (consisting of prion sequence 108-144). The amyloids were generated using these peptides in appropriate conditions and were taken to explore the degradation capacity of NK. The results showed a reduction in fluorescence emission at 487 nm of Thioflavin-T (ThT) dye (a dye that specifically binds to amyloids), negative ellipticity at 218 nm in Circular dichroism (CD) spectrum, and less fibril morphology in electron microscopic observation. Overall, this study explored the potential of NK for the degradation of amyloid at a temperature of 40 °C and pH 7 [12]. NK not only plays a role in fibrin degradation but also enhances the release of tissue plasminogen activators from cells to break down the fibrin [124].
Anti-inflammatory effect
In recent studies, an anti-inflammatory agent, like NK, is effective in suppressing various inflammatory diseases and oxidative stress (ROS). Wu et al. demonstrated that NK can suppress LPS-induced NOX2 activation and the TLR4 pathway to reduce ROS and proinflammatory mediators [125]. Moreover, long-term use of NK has shown promising results in ameliorating chronic colitis and inhibiting colonic inflammation by regulating gut microbiota and suppressing the TrP metabolism [126]. Additionally, NK exhibits an inhibitory effect against retinal neovascularization and can alleviate neuroinflammation by inducing proinflammatory microglia into an anti-inflammatory phenotype through the Nrf2/HO-1 pathway [127]. Another study by Elbakry et al. suggested that NK can protect against neurotoxicity and attenuate neuroinflammatory activity induced by certain substances. Notably, most clinical studies have indicated that short-term and low-dosage administration of NK did not induce toxicity [128].
Nattokinase’s role in other disease
Research has identified natural anticancer compounds such as phytoestroprotease inhibitors, flavonoids, and phytic acids in soybeans (natto). Furthermore, studies have shown that fermented natto contains higher levels of anticancer compounds compared to the raw material. According to Chou et al. [129], both natto freeze drying and natto water extract have been found to trigger cell autophagy in a dose-dependent manner. Hypertension stands as the most prevalent chronic illness and a significant risk factor for cardiovascular disease. NK from Ruditapes philippinarum decreased blood pressure in hypertensive rats by minimizing intestinal bacterial diseases, providing an effective defense against hypertension-induced vascular and cardiac damage [130]. Furthermore, studies indicated that oral administration of NK has decreased systolic and diastolic blood pressure by cleaving fibrinogen in plasma. Moreover, as per Keziah, NK exhibited notable angiotensin-converting enzyme inhibitor activity, achieving 87.45 % in an in vitro coagulation lysis assay. These findings suggest that NK plays a significant role in preventing and treating hypertension [131]. Similar findings were also demonstrated by another enzyme SP [132].
Lumbrokinase and serratiopeptidase in amyloid degradation
Prion protein PrPC is a cellular protein post-translationally modified into a transmissible, prion-associated protein known as PrPSc. As a result of conformational changes in the PrP 106-126 region of the PrPSc peptide, fibrillation is initiated in this protein and thus, PrP 106-126 can be used for studying prion peptide amyloidosis. There is a possibility that any agent capable of destabilizing or disintegrating such proteins can be used as a potential drug candidate for the treatment of prion diseases. The activity of LK and SP was in vitro tested against PrP 106-126 amyloids, which was measured against NK, the standard amyloid degrading enzyme. Based on ThT fluorescence assay results, both LK and SP inhibited PrP 106-126 amyloid formation. Furthermore, after incubation of prion amyloids with LK and SP, fibril sizes were also found to be smaller at different time intervals using dynamic light scattering. Furthermore, the molecular dynamics simulation revealed that PrP 106-126 had a high affinity for LK and SP. According to the study, SP and LK have the potential to disintegrate PrP 106-126 amyloids and improve the viability of the PC12 cells [133]. A similar result was also found for the amyloid β 1-42, the protein that gets misfolded and deposited in the brain in case of Alzheimer’s disease [134]. In vitro studies of the degradation of insulin amyloid using LK and SP and the results were promising [132,135]. When the study was extrapolated in an animal model, where we induced insulin amyloid mass, after repeated subcutaneous injection of insulin amyloids, we could observe that the insulin amyloid formation was retarded and the amyloids were degraded using both LK and SP [136].
Other natural compounds reported as amyloid inhibitors
Natural compounds with anti-amyloid potential are always more beneficial than synthetic ones as they are routinely consumed. Various polyphenols like epigallocatechin-3-gallate (EGCG), resveratrol and curcumin are reported for anti-amyloid potential and progressed for clinical trials. To date, 72 natural compounds are reported for the inhibition of amyloids and 44 compounds belong to a phenolic group, 4 anthraquinones, 16 flavonoids, 13 alakoloids (includes 3 pyridines, 2 porphyrins, and 3 indoles), terpenes, and steroids. Cyclodextrin, squalamine, vitamin A, hematin, rifampicin, and scyllo-inositol have anti-amyloid activity shown under in vitro conditions [137,138]. There are several agents reported as amyloid inhibitors, some of them are in clinical trials, as shown inTable 2 [139].
| Stages | Agent | Agent mechanism class | Mechanism of action | Therapeutic purpose | ClinicalTrials.gov ID [139] | Ref. |
|---|---|---|---|---|---|---|
| Phase I | Aducanumab | Anti-amyloid | Monoclonal antibody | Remove amyloid (DMT) | NCT01677572 | [140] |
| Crenezumab | Anti-amyloid | Monoclonal antibody | Remove amyloid (DMT) | NCT02353598 | [141] | |
| KHK6640 | Anti-amyloid | Anti- Aβ peptide antibody | Remove amyloid (DMT) | NCT03093519 | [142] | |
| LuAF20513 | Anti-amyloid | Polyclonal antibody | Remove amyloid (DMT) | NCT02388152 | [143] | |
| LY3002813 | Anti-amyloid | Monoclonal antibody | Remove amyloid (DMT) | NCT02624778 | [144] | |
| Phase II | BAN2401 | Anti-amyloid | Monoclonal antibody | Remove amyloid (DMT) | NCT017673311 | [145] |
| E2609 | Anti-amyloid | BACE inhibitor | Reduce amyloid production (DMT) | NCT02322021 | [146] | |
| Octagam 10 % | Anti-amyloid | 10 % human normal immunoglobulin | Remove amyloid (DMT) | NCT03319810 | [147] | |
| UB-311 | Anti-amyloid | Active immunotherapy | Reduce amyloid (DMT) | NCT0255180 | [148] | |
| Phase III | CNP520 | Anti-amyloid | BACE inhibitor | Reduce amyloid production (DMT) | NCT03131453 | [149] |
| Gantenerumab | Anti-amyloid | Monoclonal antibody | Remove amyloid (DMT) | NCT02051608 | [150] | |
| MK-8931 (verubecestat) | Anti-amyloid | BACE inhibitor | Reduce amyloid production (DMT) | NCT01953601 | [151] | |
| Solanezumab | Anti-amyloid | Monoclonal antibody | Remove amyloid (DMT) | NCT02008357 | [152] |
Further research is warranted to determine many more drugs that can cross the blood-brain barrier and effectively degrade the amyloids responsible for various neurodegenerative disorders.
Conclusion
Biotechnology has become highly versatile and has merged with other technologies to deliver high-throughput products. Its amalgamation with bionanotechnology has created tremendous demand in the biomedical field. Enzymes from natural sources have been consumed in different forms worldwide for a long time, owing to their health benefits. Among the various serine proteases, SP, LK, KER, and NK have shown their potential in inhibiting the amyloid fibril formation responsible for different neurodegenerative disorders, such as Alzheimer’s disease, Prion disease, and type II diabetes. Early diagnosis of amyloidosis permits for effectively improved organ function. Moreover, if these natural inhibitors successfully complete clinical trials, outcomes for those suffering from amyloidosis can be improved, and serine protease enzymes may be used to improve quality of life in the early stages of the disease. In this mini review, we have discussed these serine proteases and other natural compounds that can be used to manage neurodegenerative diseases. These studies concluded that KER can be used to remove prion infections from surgical instruments and other surfaces contaminated with prions because KER is not consumable. On the other hand, LK and SP can be used to degrade amyloids. Since they possess fibrinolytic activity, they can be used as coatings above surgical implants so that they can retard the clot formation at the site of implant post-surgery. Future studies are necessary to nano-formulate these enzymes to effectively cross the blood-brain barrier and get delivered to the brain for exerting their anti-amyloid activity.