Introduction
Cow milk is a nutritionally valuable food that people have been consuming for thousands of years. It contains lactose, fat, protein, ash, and numerous other biomolecules essential for growth and maintenance of consumer metabolism. Caseins and whey proteins constitute more than 95 % of bovine milk proteins. Caseins belong to major proteins in bovine milk, which can be divided into four forms: alpha s1 casein (39-46 % of total caseins), alpha s2 casein (8-11%), beta-casein (25-35 %), and kappa casein (8-15 %; Roginski, 2003). Members of a multi-gene family, which are found on bovine chromosome 6, encode them and most casein variants have been identified at the protein and DNA levels (Rijnkels, 2002). Genetic polymorphism of milk proteins commands a great interest, due to its relationships with milk production traits, composition, quality and other important traits. Beta-casein is the second most abundant protein, consisting of 209 amino acids and has excellent nutritional balance. Beta-casein gene (CSN2) is 8.5 kb in length, it contains five exons and eight introns (Bonsing et al., 1988), and has 12 known genetic variants (A1, A2, A3, B, C, D, E, F, G, H1, H2, I; Farrell et al., 2004). The occurrence of variants CSN2 is based on nucleotide exchanges located in exon VII (A1, A2, A3, B, F, G, H1, H2, I), exon VI (C, E) and exon IV (D) (Cui et al., 2011). The most common A1 and A2 variants of beta-casein differ at amino acid position 67 with histidine (CAT) in A1 and proline (CCT) in A2 milk, as a result of single nucleotide difference in the sequence of exon VII bovine CSN2 gene on the position 8101 (McLachlan, 2006; GenBank M55158). Therefore, "A1 milk" (histidine at 67 position) and "A2 milk" (proline at 67 position) can be distinguished. Kamiński et al. (2007) observed that A1 variant is most frequent in the Holstein-Friesian (0.310-0.660) (cattle) breed. Some researchers suggest a certain association of beta-casein genotypes (allele-variants) with the milk yield (Ikonen et al., 2001; Micinski et al., 2006; Kraus et al., 2016), protein content (Cardak, 2005; Micinski et al., 2006; Bonfatti et al., 2010) and fat content (Chung et al., 1996; Ikonen et al., 1999; Cardak, 2005; Oleński et al., 2012). Ikonen et al. (1999) as well as Oleński et al. (2012) observed a negative relation of allele A2 on milk fat content. Kraus et al. (2016) concluded that cows of A2A2 genotype, in comparison with other two (A1A1, A1A2), had a tendency towards higher milk yield. Ikonen et al. (2001) indicated that CSN2 variants might be useful in the direct selection programs for improving milk and fat yield although Ozdemir et al. (2018) based on meta-analysis observed that the CSN2 gene is not useful as a routinely marker for the improvement of milk production traits.
The protein fraction composition of beta-casein has become of special interest due to a possible relationship between "A1 milk" and the consumers’ health. Through the incomplete digestion of A1 beta-casein in the small intestine, it releases a bioactive peptide beta-casomorphin-7 (BCM-7) which has a cytomodulatory effect and is a risk factor for human health as it can potentially affect numerous opioid receptors in the nervous, endocrine and immune system (Svenberg et al., 1985; Elliott, 1992; Jinsmaa and Yoshikawa, 1999; Meisel, 2001). Many authors observed the impact of "A1 milk" consumption on cardiovascular diseases, sudden infant death syndrome, ischemic heart disease, type I diabetes, schizophrenia, autism and arteriosclerosis (Elliott et al., 1999; Virtanen et al., 2000; McLachlan, 2001; Monetini et al., 2002; Laugesen and Elliott, 2003; Sun et al., 2003; Tailford et al., 2003; Truswell, 2005; Woodford, 2006; Bell et al., 2006; Sodhi et al., 2012). Ho et al. (2014) observed a positive association between abdominal pain and stool consistency on the A1 diet. Jianqin et al. (2016) indicated that the consumption of milk, containing A1 beta-casein, worsened gastrointestinal symptoms, increased gastrointestinal transit time, worsening symptoms of post-dairy digestive discomfort, and decreased cognitive processing speed and accuracy in subjects with self-reported lactose intolerance. He et al. (2017) also reported that the consumption of milk containing A2 beta-casein reduced acute gastrointestinal symptoms in people who were lactose intolerant. However, a different conclusion can be drawn from the scientific community. Truswell (2005) claims in his review paper that no convincing or probable evidence was found, that the "A1 milk" is a factor causing type I diabetes and coronary heart disease. The same author indicates that the evidence relating autism and schizophrenia to "A1 or A2 milk" is rather speculative. In the scientific report elaborated by EFSA (2009), an extensive review of scientific literature on A1/A2 milk was made. They observe no cause-effect relationship between the oral intake of BCM-7 or related peptides and etiology of any suggested non-communicable diseases. Research indicates that there is still a scientific controversy about A1/A2 milk on consumers’ health, but these controversies have stimulated public interest on these issues.
An interest in beta-casein polymorphism is present with producers and consumers. In times of a low market price of raw milk, smaller and traditional milk producers seek to increase profitability by adding certain market "bonuses" to the product. One of the possibilities is the production of "A2 milk”. This possibility was recognized by breeders of local cattle breeds trying to achieve economic reaffirmation of these breeds as one of the possibilities to increase their competitiveness. However, there is a potential risk of losing a genetic variability when favouring one genetic variant in a selection. Therefore, the aim of this study was to determine the distribution of A1 and A2 beta-casein polymorphisms in conventional and local cattle breeds in Croatia. Further goal was also to compare the obtained results in the context of previous studies of beta-casein polymorphism in cattle breeds and study the potential of milk production.
Materials and methods
In this research three conventional (milk production) and three local breeds (breeds in the program of protection and search for ways of better economic reaffirmation) in Croatia were included: as follows Holstein (HOL; N=60), Simmental (SIM; N=60), Brown Swiss (BSW; N=60), Busa cattle (BC; N=49), Slavonian-Syrmian Podolian cattle (SSPC; N=49) and Istrian cattle (IC; N=43). Data from milk yield and chemical composition of conventional breeds (HOL, SIM, and BSW) in the first three lactations were provided from the central database of the Croatian Agricultural Agency.
Hair samples of the SIM breed and the SSPC were collected in the Zagreb and Koprivnica-Križevci Counties, for the HOL breed in the Osijek-Baranja County, for BSW and IC in the Istria County, and for the BC in Dubrovnik-Neretva and Lika-Senj Counties. After collecting, tissue samples were stored at -20 °C until DNA isolation was performed using Sigma-AldrichTM GenElute Mammalian Genomic DNA Miniprep Kit (www.sigma-aldrich.com). Isolated DNA was stored at -20 °C until the genotyping procedure was performed. The polymerase chain reaction (PCR) was used to amplify target codogenic DNA regions of the genes responsible for the occurrence of the beta-casein (CSN2) polymorphism. Initial oligonucleotide sequences (Forward: 5´- CCT TCT TTC CAG GAT GAA CTC CAG G- 3´ and Reverse 5´ - GAG TAA GAG GAG GGA TGT TTT GTG GGA GGC TCT- 3´; McLachlan, 2006) were used to amplify the codogenic regions of the CSN2 gene. Amplification of the 121 bp codogenic sequence was performed in a 13.8 μL reaction mixture (5.4 μL MiliQ water, 7.5 μl EmeraldAmp GT PCR Master Mix, 0.45 μL of each primer and 1.2 μL DNA). The polymerase chain reaction was performed on a Thermal Cycler MJ Research PTC 100 according to a temperature program to which, after initial denaturation (98 °C/3 min), 35 cycles of DNA fragment amplification followed (98 °C/10 sec, 58 °C/30 sec, 72 °C/50 sec) and final extension 72 °C/5 min). A quality control of the PCR reaction was performed on a 1 % agarose gel after staining with StainIN Green Nucleic Acid.
The identification of A1 and A2 allelic variants of CSN2 was performed according to Miluchová et al. (2014). After the amplification, the PCR product was digested by restriction endonuclease DdeI for 4 h/37 °C in the prepared reaction mixture (0.40 μL dH2O, 10 μL PCR product, 0.10 μL DdeI enzyme). Restriction enzyme splitting results were read directly on a 3 % agarose gel after electrophoresis (85V/40min; Figure 1).
The determination of allelic and genotype frequencies and a Hardy-Weinberg equilibrium exact test were estimated using the software GenPop V4.3 (Raymond and Rousset, 1995, Rousset, 2008). Testing the excess and deficiency of heterozygotes and the estimation of F IS index were carried out using the same program. The observed and the expected heterozygosity was computed with Genetix v4.05.2 (Belkhir et al., 2004). The effect of CSN2 genotype on milk yield and chemical composition of milk in a standard lactation interval was determined using the linear models (GLM procedure, SAS STAT, V8, 1999) with included fixed effects of CSN2 genotype, while the breed was included as linear covariate. The following statistical model was used to estimate effects on milk yield and composition: Y ijk = μ + βCSN2 i + B j + e ijk , where Y ijk is the observed value of phenotype, μ is the general mean, βCSN2 i is the effect of the CSN2 genotype ( i = A1A1, A1A2, A2A2), B j is the effect of the breeds ( j = HOL, SIM, BSW), and e ijk is the random residual effect.
Results and discussion
Visualization by 3 % agarose-gel electrophoresis of DdeI digestion of the CSN2 PCR product is shown in Figure 1. CSN2 genotypes are determined based on the size of the DNA fragment: A1A1 (121 bp), A1A2 (121, 86, 35 bp) and A2A2 (86, 35 bp).
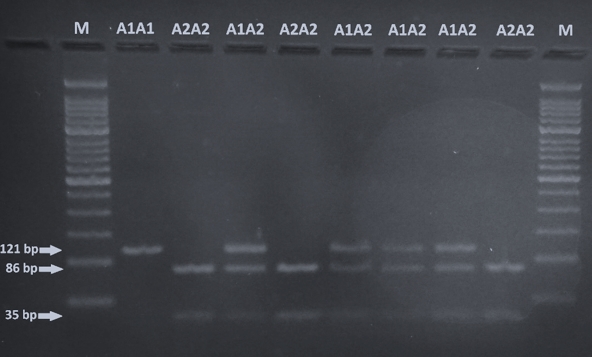
Figure 1. Gel electrophoresis of the amplified 121 bps fragment of CSN2 gene digested with DdeI (M, 50 bp ladder; A1A1 - 121 bp, A1A2 - 121, 86, 35 bp, A2A2 - 35 bp)
Table 1 summarizes the genetic diversity parameters of the CSN2 gene in six cattle breeds. Allele A2 was the most frequent in all breeds ranging from 0.650 (HOL) to 0.758 (SIM). Contrary, the frequency of the A1 allele was on average 1.9 times smaller ranging from 0.242 (SIM) to 0.350 (HOL). The predominant genotype frequency was A2A2 in local (IC, SSPC) and conventional (SIM) cattle breeds with frequencies >53.06 %. A frequency of heterozygous genotype A1A2 was observed in one local breed (BC) and one conventional breed (BSW), thus BSW having higher frequencies (51.67 % vs. 44.90 %). The lowest frequency was determined for the A1A1 genotype ranging from 3.33 (SIM) to 18.37 (SSPC).
Table 1. Estimates of allele and genotype frequency, observed (H O) and expected heterozygosity (H E), fixation index (F IS) with level of significance for CSN2 gene in six cattle populations
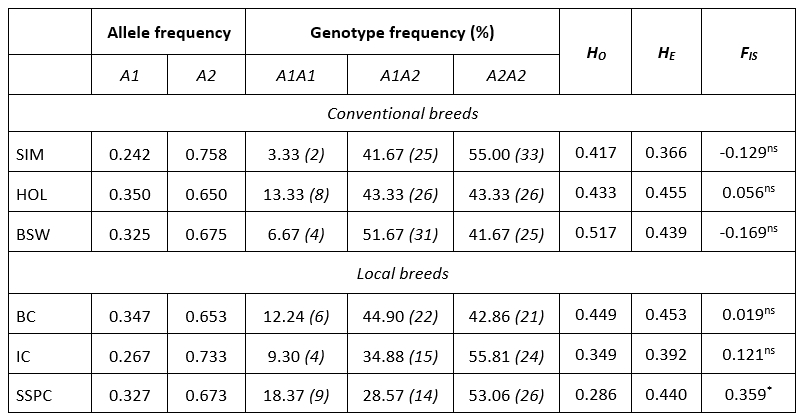
ns: non-significant; *: P<0.05; number of animals is indicated in the brackets
The obtained CSN2 allele frequency results for conventional breeds (SIM, HOL and BSW) are comparable and were within a range of previous researches presented in Figure 2. The observed dominance of the A2 allelic variant of the CSN2 gene in the HOL population coincided with most previous studies, except for the population in Australia, the United States, the United Kingdom, and Ireland (McLean et al., 1984; O’Hara, 1995; Jann et al., 2004). A dominance of the A2 allelic variant (fA2 = 0.68) was also observed in the population of BSW cattle as was in related populations (Van Eenennaam and Medrano, 1991; Boetcher et al., 2004). In the SIM breed population, A2 allelic variant of the CSN2 gene is dominant (fA2 = 0.758), which corresponding to the results from previous studies (Figure 2). Compared to the previously determined frequencies of allelic variants of the CSN2 gene (Curik et al., 1997), an increase of the A2 allelic variant by 0.13 (0.63 0.76) was observed in the Simmental population in Croatia. The observed change in the frequencies of allelic variants is partly due to the limited introduction of the Red Holstein genome into the Simmental population at the beginning of the 21 st century, primarily to improve milking and milk characteristics.
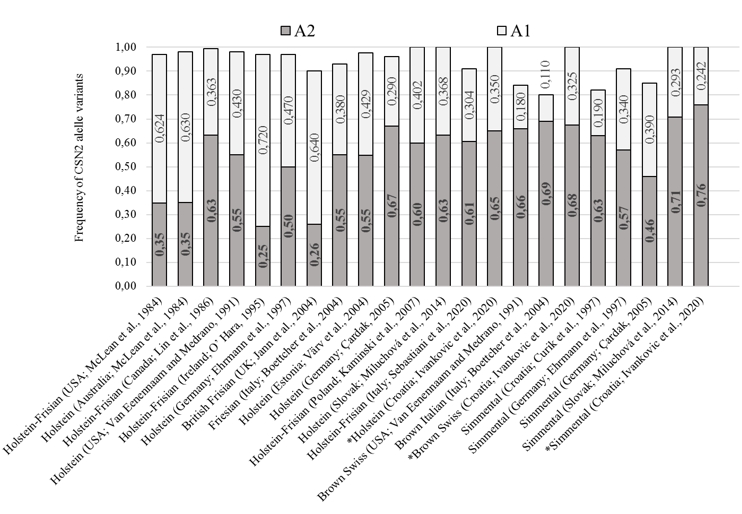
Figure 2. Share of A1 and A2 CSN2 gene variants in Holstein, Brown Swiss and Simmental populations (different literature sources). Results from this research are marked with asterisks (*)
In local populations, A2 allelic variant of the CSN2 gene dominates, which is expected given the observations of numerous previous studies on the dominance of the A2 variant in local cattle breeds. In the population of IC, the presence of the A2 variant of CSN2 gene increased by 0.23 (0.51 vs. 0.73) compared to the previous research (Jann et al., 2004). Baranyi et al. (1993) also observed the dominance of the A2 variant of the CSN2 gene in the Hungarian Gray cattle population that is phylogenetically related to SSPC (fA2 = 0.75). Considering the obtained results, we conclude that local breeds have a predisposition for the production of desirable "A2 milk" through herd formation consisting of homozygous A2 individuals. Most certainly, adequate care should be taken not to disrupt genetic variability as a primary goal of breeding local endangered breeds by favouring the A2 allele variant. If selection supporting the SCN2 A2 variant is to be carried out within local cattle breeds, it is necessary to preserve part of the population that will serve as a nucleus (with less represented CSN2 gene variants and rarer lines).
The observed heterozygosity ( H O ) ranged from 0.286 in SSPC to 0.517 in BSW while, the expected heterozygosity ( H E ) ranged from 0.366 in SIM to 0.455 in HOL population. In general, compared to local, conventional breeds capture a higher degree of genetic variability ( H O 0.454 : 0.427). Higher values for H E in Estonian dairy cattle (0.526) were reported by Värv et al (2009). In the research of Miluchovà et al. (2014) results of the genetic structure of local Slovak spotted cattle was similar to those for IC and SSPC, but rather small for BC. The same author reported heterozygosity values for Holstein breed ( H O = 0.460, H E = 0.470) which were close to the results from this research for HOL breed (Table 1). Based on the exact probability test, five breeds were in Hardy-Weinberg equilibrium, except SSPC. The extent of divergence of the F IS values from zero was notable (0.121 for IC, -0.129 for SIM and -0.169 for BSW) but in all cases significant heterozygote deficiency or excess was not established in the populations. An exception was the SSPC population which showed a deficit of heterozygotes of 36 % (p = 0.014). In recent past, the population of SSPC experienced the bottleneck effect (Ramljak et al., 2011; Ramljak et al., 2018) and very likely the founder effect (personal communication). These evolutionary processes accompanied by consequent inbreeding due to small population size (in 2018. 227 cows and 13 bulls; MA, 2019), resulted in such high F IS value. Värv et al. (2009) reported a higher, but not significant value of inbreeding coefficient in three dairy breeds form Estonia. In conventional breeds, F IS values were negative or, in case of HOL positive without significance, suggesting well-run management and breeding practice. In those populations, mating is conducted via planned schemes applying artificial insemination what support genetic diversity within populations, but also has influence for economic traits.
An association of allelic gene variants with the production characteristics of dairy cows is an important motive for their inclusion in selection programs. Ikonen et al. (2001) indicated that CSN2 variants might be useful in selection programs, although Ozdemir et al. (2018) did not observe its usability. Consumers’ interest in health aspects of milk protein further motivates farmers to selectively propagate desirable allelic variants. Indicators of milk yield and milk chemical composition in the first three lactations with respect to A1 and A2 CSN2 genotypes are shown in Table 2. Higher lactation production was observed in the A2A2 genotype in the first three lactations, but only the observed difference in the second lactation was significant (A2A2 vs. A1A1; p<0.05). The observed positive effect of A2A2 CSN2 genotype on lactation production has been observed in previous studies (Ikonen et al., 2001; Micinski et al., 2006; Kraus et al., 2016).
Table 2. Indicators of milk production and chemical composition in the first three lactations by CSN2 gene genotypes (LSMEAN±SE)
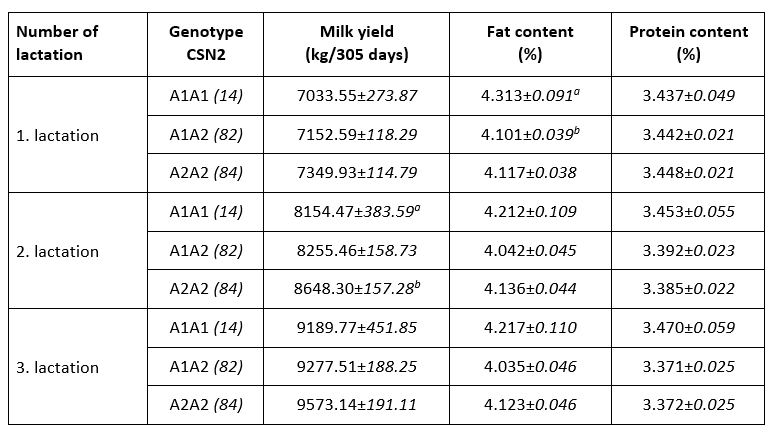
p<0.05
A higher proportion of milk fat was observed in the milk of cows of A1A1 CSN2 genotype, but only the difference in the first lactation was significant (A1A1 vs. A1A2; p<0.05). Earlier studies (Chung et al., 1996; Ikonen et al., 1999; Cardak, 2005; Oleński et al., 2012) observed a positive relation of allele A1 on milk fat content what is in line with recent observations. No effect of genotypes of CSN2 gene on milk protein content during three lactations was noted, which is consistent with previous research (Van Eenennaam and Medrano, 1991; Micinski et al., 2006). The conducted research indicates the existence of certain weak connections of allelic variants of the CSN2 gene with milk production, the share of milk fat and milk proteins, which supports the dilemma from earlier published observations. The observed relationships of CSN2 genotypes and production characteristics can be integrated into selection programs of conventional breeds to increase lactation production or milk fat content, taking into account the balance of these characteristics and consumer preferences ("A2 milk"). Favouring the A2 allelic variant of the CSN2 gene in the selection of local (and endangered) cattle breeds is one possibility for their reaffirmation and increased competitiveness through production of milk with added value.
Conclusions
Local (BC, IC, SSPC) and conventional breeds (SIM, HOL, BSW) exhibit superiority of allele A2. In addition, the favourable genotype A2A2 is present in IC, SSPC, SIM and HOL breeds. Five breeds maintain high level of genetic diversity at CSN2 locus except SSPC. Higher milk production was determined in conventional breeds of A2A2 genotype while higher fat content was obtained by A1A1 genotype. These findings indicate the genetic potential of conventional and especially local breeds for the production of CSN2 A2A2 milk as a functional food (might lower the risks associated with gastrointestinal and coronary diseases, diabetes, allergies, etc). The oriented production of A2A2 milk could represent an added value to local breeds ensuring its sustainability and supporting biodiversity by opening the door for finding a new role in the future for these breeds.
Genetski polimorfizam i utjecaj na proizvodnju mlijeka CSN2 gena u konvencionalnim i lokalnim pasminama goveda u Hrvatskoj
Sažetak
Tijekom protekla tri desetljeća interes znanstvene i ukupne javnosti ( potrošača) potaknut je studijama u kojima je zapažen negativan učinak konzumacije mlijeka u kojem je A1 varijanta beta-kazeina na zdravlje potrošača. Proizvodnja "A2 mlijeka" jedan je od načina gospodarskog povećanja kompetitivnosti manjih i srednjih mliječnih farmi. Uzgajivači ugroženih lokalnih pasmina također su zainteresirani za reafirmaciju kroz proizvodnju "A2 mlijeka". Cilj istraživanja je utvrditi frekvencije A1 i A2 varijanti beta-kazeina u konvencionalnim i lokalnim pasminama goveda u Hrvatskoj, te njihovu povezanost s proizvodnim pokazateljima konvencionalnih pasmina u prve tri laktacije. Genomska DNK izolirana korištenjem komercijalnog kita za izolaciju upotrijebljena je za PCR‑RFLP determinaciju CSN2 genotipova. Podaci o mliječnosti i kemijskom sastavu mlijeka u promatranom uzorku preuzeti su iz središnje baze podataka. Utvrđena je dominantna zastupljenost A2 varijante beta-kazeina u istraženim pasminama goveda (0,650-0,758) te povećanje frekvencije A2 beta-kazeina u populaciji simentalskog i istarskog goveda. Zapažena je povezanost A2A2 i A1A1 genotipa beta-kazeina s laktacijskom proizvodnjom i udjelom mliječne masti u prvoj i drugoj laktaciji (p<0,05). Dominacija A2 alelne varijante beta-kazeina čini pogodnim istražene konvencionalne i lokalne pasmine za proizvodnju "A2 mlijeka". Uzgojno protežiranje A2 alelne varijante beta‑kazeina unutar lokalnih pasmina goveda treba provoditi pažljivo kako ne bi izgubili dio zatečene genetske varijabilnosti.
Ključneriječi: beta-kazein; pasmine goveda; polimorfizam; A2 mlijeko
