1. Introduction
The enzyme endo-1,4-xylanase, or generally xylanase, has been attracting increasing attention in recent decades due to its possible application in lignocellulosic biorefinery (Liao et al., 2022) which is one of the key enabling strategy of the circular bioeconomy (Schmitz et al., 2022). It is used in feed production, food industry (bakery (Bajaj and Manhas, 2012), fruit juice (Kaushal et al., 2021) and beer clarification (Walia et al., 2017), leather processing, pulp and paper bleaching, and production of biofuels from lignocellulosic biomass (Uday et al., 2016; Kumar and Shukla, 2016).
A large number of microorganisms such as bacteria, fungi and yeasts are capable of producing xylanase. Today, xylanase is commercially obtained mostly from filamentous fungi, as their cultivation is the easiest way to produce large quantities of xylanase (Bhardwaj et al., 2019). In order to meet industrial needs for stable and active enzymes, an intensive search for new xylanase sources has been carried out in recent decades. Once a suitable xylanase-producing microorganism is found and the enzyme is isolated, one of the major limiting factors in xylanase characterization and further proper use is the lack of a standard method for determining its enzymatic activity. This is primarily related to the problem of aligning the results of xylanase-catalyzed processes derived from different sources. Different researchers (Moran-Aguilar et al., 2021; Hassan et al., 2021; Jain et al., 2020; Balderas Hernández et al., 2021; Kumar et al., 2017; Abdella et al., 2021; Kim et al., 2021) use different methods to determine the activity of the xylanase, and therefore great effort and resources are invested to define a standard method.
The most common method for determining the xylanase activity is the so-called DNS (3,5-dinitrosalicylic acid) spectrophotometric method. The method was first proposed by Bailey et al. (1992), and it was based on the measurement of the visible absorbance of the product (3-amino-5-nitrosalicylic acid) formation. It is a rapid method that can replace time-consuming methods such as Liquid Chromatography (HPLC) coupled to Mass Spectrometry, Evaporative Light Scattering, Refraction index or Standard four-potential-pulse amperometric detectors (Wood et al., 2012; Li et al., 2021). Over the years, it has been found that the method has one significant drawback, and that is the loss of some of the reduced sugar during analysis. This has already been found in several studies (Summer and Grahram, 1921; Summer and Noback, 1924; Summer, 1925; Miller, 1959) and the composition of the reagent responsible for this phenomenon has been changed and the method modified (Miller, 1959). In addition, the concentration of the compounds, the boiling time, and the dilution ratios were additionally optimized. Even though there were still problems to be solved, the method has remained essentially the same and is also recommended by the International Union of Pure and Applied Chemistry (IUPAC) for the determination of reducing sugars (Saqib and Whithey, 2011). The main problems are related to chemical interferences and the fact that many factors can affect the formation of 3-amino-5-nitrorsalicylic acids, such as the presence of various sugars (Bell et al., 1951) and sugar degradation products, i.e. furfural (Deshavath et al., 2020), the presence of aldehydes, amino acids (Teixeira et al., 2012), etc. In addition, the presence of some salts in the buffer such as calcium chloride, manganese sulphate and cobalt can increase the absorption of the DNS method. On the other hand, some agents such as EDTA may have the opposite effect (Teixeira et al., 2012). In addition, according to Wood et al. (2012) reaction volume, incubation time and the measurement of wavelengths can also affect the DNS method. Nevertheless, the basic settings of the method have not been re-evaluated in recent studies, although it is known that the concentration of the substrate, the enzyme concentration, the working volume of the assay, etc. can affect the calculation of xylanase activity.
The aim of this study is to modify/standardise the existing DNS method for the determination of xylanase activity by analyzing the effects of various DNS reagents, various substrates and their concentrations, incubation time and buffers on the assay. Potential problems were revealed and new solutions were proposed. As a result of this research, a modified method for determining the xylanase activity was proposed.
2. Materials and methods
2.1. Chemicals
3,5-dinitrosalicylic acid and phenol were purchased from Fluka (China), arabinoxylan from Megazyme (Ireland), citric acid from VWR Chemicals (Belgium), D(+)-xylose from BDH Prolabo (United Kingdom), bovine serum Albumin (BSA) and xylan from beechwood from Sigma-Aldrich (Austria), xylan from corn kernel from TCI America (Japan), xylanase from Aspergillus niger (S.A. = 300 U/g and S.A. = 3400 U/g), xylanase from Bacillus SMB (S.A. = 586 U/g), xylanase from Trichoderma reesei (S.A. = 300 U/g) from AB Enzymes (Germany), and xylanase from Trichoderma longibrachiatum (S.A. = 1000 U/g) were purchased from Biocat (USA). Sodium dihydrogen phosphate dihydrate, sodium metabisulfite, sodium-potassium-tartrate-tetrahydrate and sodium sulphite (anhydrous) were obtained from Kemika (Croatia). Sodium hydroxide was purchased from Gram Mol (Croatia), and potato dextrose agar was purchased from Biolife (Italy). The chemicals were of analytical grade and were used without further purification.
Sugar beet pulp pellets were obtained from the factory for processing sugar beet production and sugar processing, Sladorana Ltd. Županja, Croatia.
Trametes versicolor TV-6 was obtained from the National Institute of Chemistry, Slovenia.
2.2. Methods
2.2.1. DNS reagent preparation
DNS reagent was prepared according to three methods:
DNS reagent I was prepared according to Gusakov et al. (2011). 0.02 g/mL DNS solution was prepared by dissolving 3,5-dinitrosalicylic acid in 0.5 mol/L sodium hydroxide solution at 70 °C. After the 3,5-dinitrosalicylic acid was completely dissolved, 60 g of sodium potassium tartrate (Rochelle salt) was added gradually until it was completely dissolved. The prepared reagent was stored in a dark reagent bottle until use.
DNS reagent II was prepared according to the method described by Bailey et al. (1992) by dissolving (mass fractions): 1% 3,5-dinitrosalicylic acid, 0.2% phenol, 0.05% sodium sulphite, 1% sodium hydroxide and 97.75% distilled water. The prepared reagent was stored in a dark reagent bottle until use.
DNS reagent III was prepared according to the method described by Sumner and Sisler (1944). Briefly, 1 g of NaOH was added to 70 mL of distilled water with constant stirring on a magnetic stirrer. After dissolving NaOH, the following chemicals were added: 30 g sodium potassium tartrate, 1.06 g 3,5-dinitrosalicylic acid, 0.83 g sodium metabisulfite and, 0.2 g phenol. The total volume was then adjusted to 100 mL with distilled water. The prepared reagent was stored in a dark reagent bottle until use.
2.2.2. Substrate solutions
Arabinoxylan, xylan from beechwood and xylan from corn kernel (1 g) were added to 80 mL of 0.05 mol/L Na-citrate buffer, with a pH value of 5.3 and were dissolved by stirring on a magnetic stirrer at 60 °C. After cooling down to 25 °C, the volume was adjusted to 100 mL and the prepared solutions were stored at 25 °C for no more than one week.
2.2.3. Preparation of xylanase stock solutions
Stock solutions (1 mg/mL) of enzymes, xylanase from A. niger (S.A. = 300 U/g and S.A. = 3400 U/g), xylanase from Bacillus SMB (S.A. = 586 U/g), xylanase from T. reesei (S.A. = 300 U/g) and xylanase from T. longibrachiatum (S.A. = 1000 U/g), typically used in the bakery industry, were prepared by dissolving 1 mg of enzyme in 0.05 mol/L Na-citrate buffer, with a pH value of 5.3. The enzyme concentration in stock solutions was determined according to the linearized Bradford protein assay (Ernst and Zor, 2010).
2.2.4. Determination of xylanase activity
A stock solution of xylose (0.01 mol/L) was prepared in a 0.05 mol/L Na-citrate buffer with a pH value of 5.3. Xylose solutions of various concentrations (0.5 µmol/mL, 1 µmol/mL, 2 µmol/mL, 3.33 µmol/mL, 5 µmol/L and 10 µmol/mL) were prepared by diluting 0.01 mol/L xylose stock solution with 0.05 mol/L Na-citrate buffer with a pH value of 5.3. The prepared solutions were used to make the calibration curve. After preparation of the xylose solutions, 0.9 mL of xylan solution and 0.1 mL of xylose solution were mixed and heated at 50 °C for 300 s. 1.5 mL of DNS reagent was added to the mixture. The reagent and mixture were heated at 100 °C for 5 min. The mixture was then cooled to 25 °C in a water bath and the absorbance at λ = 540 nm was measured with a spectrophotometer (UV-1601, Shimadzu, Japan). To monitor the spontaneous decomposition of the substrate, a substrate blank was prepared using 0.1 mL of 0.05 mol/L Na-citrate buffer of 5.3 pH instead of 0.1 mL of xylose solution. The calibration curve was plotted as a function of the difference in absorbance of the xylose solution sample and a substrate blank sample against the known xylose concentration in the xylose solution sample (Bailay et al., 1992).
Xylanase activity (U/g) was determined by the same procedure using 0.1 mL of enzyme solution instead of xylan solution, and the activity was calculated based on the ratio of sugar concentration (determined according to the calibration curve) to incubation time (Equation 1).
Volumetric enzyme activity of 1 U represents the amount of enzyme required to oxidize 1 μmol of substrate per minute. If the dilution is rather small and/or if the sample contains a high level of reducing sugars, an enzyme blank is required. Enzyme blank was prepared by heating 0.9 mL of xylan solution to 50 °C for 300 s and then 1.5 mL of DNS reagent was added followed by 0.1 mL of enzyme solution. The mixture was heated to 100 °C for 5 min, cooled to 25 °C, and absorbance was determined at λ = 540 nm.
2.2.5. Interference of DNS reagent and xylan
The interference of DNS and selected substrates was determined by mixing 0.9 mL of xylan solution and 0.1 mL of 0.05 mol/L Na-citrate buffer of 5.3 pH at 50 °C for 300 s. The reaction was stopped by adding 1.5 mL of DNS. The resulting mixture was heated at 100 °C for 5 min. The mixture was then cooled to 25 °C in a water bath and the absorbance was measured at λ = 540 nm. The concentration of xylose released was calculated from the obtained absorbance, using a previously prepared calibration curve. Three, chemically different substrates, were compared: arabinoxylan, xylan from beechwood and xylan from the corn kernel. Structure, molecular mass and the composition of selected xylans are summarized in Table 1.
Table 1. Properties of different xylans
2.2.6. Interference of DNS reagent and xylanase
The interference of DNS and selected enzymes was determined by mixing 0.9 mL of 0.05 mol/L Na-citrate buffer of 5.3 pH and 0.1 mL of xylanase solution at 50 °C for 300 s. The reaction was stopped by adding 1.5 mL of DNS. The resulting mixture was heated at 100 °C for 5 min. The mixture was then cooled to 25 °C in a water bath and the absorbance at λ = 540 nm was measured. The concentration of the released xylose was calculated from the obtained absorbance, using a previously prepared calibration curve. The procedure was repeated with 2- and 10-fold diluted xylanase samples.
2.2.7. Determination of extinction coefficient
To determine the extinction coefficient, the absorbance of different xylose concentrations was measured. The extinction coefficient was calculated according to Beer's Law by dividing the absorbance by the concentration and the light path length.
2.2.8. Chemical hydrolysis of xylan
To determine the total amount of xylose in xylans, chemical hydrolysis was carried out. It was performed by heating a 1 % (v/v) solution of xylan with 2 % (v/v) acetic acid at 100 °C for 3 hours. Then, the concentration of released xylose was determined by the DNS assay.
2.2.9. Production of crude xylanase extract
T. versicolor TV-6 was cultured on potato dextrose agar at 27 °C for 14 days. Ground sugar beet pulp (25 g) was mixed with 50 mL of distilled water in laboratory jars and inoculated with T. versicolor TV-6 under solid-state fermentation conditions. A total of four jars were inoculated and incubated for eight days at 27 °C in an incubator (KB 115, BINDER GmbH, Germany). After eight days of solid-state fermentation, the material in jars was mixed and homogenized for the extraction of crude xylanase extract. The sample (12 g) was taken from the homogenized biomass and extracted in 120 mL of distilled water and it was then agitated on the magnetic stirrer (IKA, RCT basic) at 750 rpm for 60 minutes and centrifuged at 10000 x g for 10 minutes (Z 326 K, Hermle Labortechnik GmbH). The obtained supernatant was stored at +4 °C and -20 °C conditions and used as a crude enzyme extract for measuring xylanase activities.
2.2.10. Kinetic measurements
The initial rate method was used to estimate the parameters of the kinetic model, and the corresponding experiments were performed in batch reactors ( V = 2 mL). The reaction mixture was stirred on a laboratory shaker (600 rpm, T = 50 °C) (Digital Heating Shaking Drybath, Thermo Fisher Scientific, USA). The influence of xylan concentration on the rate of the hydrolysis reaction was studied in the range of xylan content from 0 to 2% (w/w). The slope, i.e. the reaction rate, was calculated for each mass fraction of xylan by monitoring the course of the reaction for 5 min and taking five samples (one sample per minute). Enzyme activity was calculated based on the ratio of sugar concentration and incubation time.
2.2.11. Data processing
Kinetic parameters were estimated by nonlinear regression analysis, i.e. using the simplex method and least squares method based on experimental data from kinetic measurements performed in a batch reactor. The software package SCIENTIST (MicroMath Scientist®, 3.0, MicroMath Scientific Software, Salt Lake City, UT, USA) was used for the estimation of kinetic parameters. The same software package was used to solve a mathematical model of the process consisting of kinetic and mass balances and describing the dynamic change of xylan and xylose concentrations for the reaction performed in a batch reactor.
3. Results and discussion
3.1. Reagent selection
There are several methods for preparing DNS reagents used in assays for xylanase activity measurements (Bailey et al., 1992; Gusakov et al., 2011; Sumner and Sisler, 1944). The DNS reagent is used in assays to stop the enzymatic reaction in which xylan was used as a substrate, and to determine the amount of reduced sugar (xylose), which is the basis for calculating the activity of the xylanase.
To prepare the DNS reagents, other chemicals apart from 3,5-dinitrosalicylic acid are used. This is usually sodium hydroxide, which is added to ensure alkaline conditions necessary for acid reduction. Another common component in DNS reagent is sodium potassium tartrate, which is added to stabilize the resulting colour after reduction. To increase the amount of colour formed, phenol is added. Bisulphite is added to stabilize the colour obtained in the presence of phenol (Miller, 1959). On the other hand, some researchers (Wood et al., 2012; Saqib and Whitney, 2011) reported that phenol should be avoided because of its toxicity and because phenol unnecessarily increases the colour intensity, especially in sugar-rich samples.
In this study, three different DNS reagents named DNS I, DNS II, and DNS III were prepared and compared to find the best one for determining the xylanase activity.
For the validation of the reagents, the following parameters were studied: the cost of reagent/assay, linearity (value of R2), repeatability, and selectivity (the interference of the reagents with substrate and enzyme).
The price of the reagent was calculated based on the chemical prices available online (Sigma Aldrich) and considering the cost of all chemicals needed to prepare each DNS reagent. Table 2 shows the cost of producing 1 L of the DNS reagent. It is assumed that for each measurement, 4.5 mL of reagent, which includes a blank substrate, a blank enzyme probe, and a reaction, is used resulting in approximately 222 analyses performed with 1 L of the DNS reagent.
Table 2. Cost estimation for the preparation of 1 L of the DNS reagent
As can be seen the DNS I reagent, which consists of only three components, is the most expensive. The reason is the large amount of 3,5-dinitrosalicylic acid and sodium potassium tartrate used in its preparation. On the other hand, the reagent DNS II is the cheapest. However, it should be noted that both DNS II and DNS III reagents, respectively, contain phenol, which is considered a disadvantage due to the environmental and health risks of its use.
To determine linearity for all three reagents, calibration standard curves were plotted where absorbance was the function of the concentration of xylose solutions for concentrations lower than 10 µmol/mL. Coefficients of determination were calculated based on the obtained data. It was found that all three calibration curves were linear in the xylose concentration range from 0 to 10 µmol/mL, which means that all three reagents can be used for xylose determination. The following correlation of determination was calculated: R2 (DNS I) = 0.9924, R2 (DNS II) = 0.9947 and R2 (DNS III) = 0.9199, respectively. All correlations of determination were found to be statistically significant, considering the 95% confidence interval. Shortly, the application of the reagents DNS I and DNS II exhibited an acceptable deviation from linearity, while the calibration direction for the reagent DNS III was not satisfactory.
The repeatability of the method using DNS I, DNS II and DNS III reagents was determined by measuring the enzyme activity for xylanase solutions prepared in various concentrations (0.0256, 0.0133 and, 0.00256 g/L). Xylanase from T. longibrachiatum with the activity of 1000 U/g declared by the manufacturer was selected as the reference enzyme. Xylanase activity was determined using assay and was compared with the enzyme activity reported by the manufacturer. The obtained data are shown in Table 3.
Table 3. Xylanase activity for xylanase solutions of different concentrations obtained in assay performed with three DNS reagents
From the obtained results, it can be seen that the assay performed with reagents DNS III has a low repeatability. On the other hand, acceptable repeatability was obtained with the assays where reagents DNS I and DNS II were used if the concentrations of enzyme in solution were 0.0133±0.0012 g/mL and 0.00256±0.0029 g/mL, respectively. The deviation that occurred when the enzyme was not diluted is probably due to its high concentration, i.e. high measured absorbance, which also affects the measurement error. In addition, due to the phenol present in the reagent (Miller, 1959), the enzyme activity is slightly higher than the declared one when DNS II was used. Finally, all measurements resulted in the xylanase activity that was significantly different from that reported by the manufacturer. The reason for this, as mentioned earlier, is the lack of a standard method for measuring xylanase activity, which is evident from performed assays, where different methods resulted in significantly different values of the measured activities.
To determine whether selected reagents interfere with other components (analytical selectivity (IUPAC, 2001)), the interactions of DNS reagents with different substrates and components present in the xylanase obtained from different origins were examined. In the measurement of xylanase activity, the decomposition of xylan occurs under the influence of high temperatures and the action of the reagent itself (alkaline hydrolysis). The following substrates were tested in this study: arabinoxylan, xylan from corn kernel, and xylan from beechwood. By this time, all reagents had been used for more than a week and it was found that negative values for xylanase concentrations were obtained when the reagent DNS II was used (Table 4). The reason is probably the instability of this reagent, which decomposes with time. In addition, according to Miller (1959), the colour obtained in the test is unstable when the Rochelle salt is absent from the reagent. Since all the reagents were prepared at the same time, it can be observed that the DNS II reagent is the most unstable.
Table 4. Xylose concentration in the sample after reaction with xylans originated from different sources
| Xylan origin | |||
|---|---|---|---|
| γxylose (μg/mL) | |||
| DNS I | DNS II | DNS III | |
| Arabinoxylan | 77.98±10.13 | - | 31.47±0.01 |
| Xylan from corn kernel | - | - | - |
| Xylan from beechwood | 91.07±12.37 | - | 36.76±0.02 |
When the DNS I and DNS III reagents are compared, it can be seen that the obtained sugar concentrations are low and can be considered negligible (the range of xylose concentration in the calibration curve was between 150 and 1500 µg/mL). The same effect was noted by McCleary and McGeough (2015), who compared the influence of the substrate on the colour formation. They prepared standard curves for xylan in the presence of beechwood glucuronoxylan, birchwood glucuronoxylan and wheat flour arabinoxylan, and all curves were similar. In addition, it was noticed that xylan from corn kernel is not acceptable as a substrate while absorbances greater than 2 were achieved during the determination of xylose concentrations. This phenomenon was explored in the following part of the research.
Since the DNS reagent can also react with different components in the enzyme stabilization mixture, the effect of the selected DNS reagents on the xylanase obtained from different origins ( A. niger, Bacillus SMB, T. reesei and T. longibrachiatum) was investigated. This analysis showed that all reagents that were used had the same colour intensity in the reaction, which means that there were no side reactions.
Considering all the observations, it was concluded that among the three reagents compared, the DNS I reagent is the most suitable for the DNS assay because it requires a minimum number of chemicals, the correlation of determination of the calibration curve is statistically significant, the repeatability of the method using the DNS I reagent is satisfactory, the use of phenol is avoided, the reagent is stable and substrates-enzyme interactions are negligible.
3.2. Substrate selection
Selection of a suitable substrate for the xylanase assay is one of the crucial steps. The substrate has to be sufficiently pure and well-defined (McCleary and McGeough, 2015). Until recently, the most commonly used substrate was xylan (4- O-methyl glucuronoxylan) from birchwood. Unfortunately, xylan from birchwood is now no longer commercially available, so new substrates have been investigated. In the present study, three substrates, arabinoxylan, xylan from corn kernel, and xylan from beechwood, were selected and compared. The properties of each selected xylan are listed in Table 1. As can be seen, arabinoxylan is highly branched and consists of xylose and arabinose (62:38). Xylan from beechwood contains 85.6% xylose while the rest are glucuronic acid and sugars. According to the manufacturer, xylan from corn kernel consists of >75% xylose and the rest are arabinose and glucuronic acid. When the enzyme assay was performed with xylan from corn as a substrate, absorbance greater than 2 was measured and absorbance values were outside of the calibration range. The first assumption was that too high concentration of the enzyme was used in the measurement, resulting in intense colouring of the solution. Therefore, further experiments were performed where the enzyme stock solution was diluted 10, 100, 1000 and 10000 fold. In these experiments, the substrate concentration was constant (1%), and the incubation time (5 minutes according to the method) was shortened to 1 minute. Despite the lower enzyme concentration and shorter incubation time, the absorbance of samples after incubation was still outside the calibration range. The method was further developed by changing the pH of the buffer and the incubation temperature. The pH of the Na-acetate buffer was changed to 5, while the incubation temperature was decreased from 50 to 25 °C to slow down the reaction. Still, the absorbance of the resulting samples was higher than 2, and no difference was observed between the absorbance of the sample after incubation and the absorbance of the blank sample.
Since the absorbance of the blank sample was also extremely high, it was assumed that the selected substrate was not pure enough for usage in reducing sugar-based assay. The selected substrate probably contains a high amount of sugar that made even blank samples after incubation too dark. Contrary, when arabinoxylan and xylan from beechwood were used, the absorbance achieved was within the range defined by the calibration. Therefore, it was concluded that these two substrates can be used in further research.
3.3. Buffer selection
To ensure acidic conditions, citric acid buffer is used in the DNS assay. In the original Bailey’s method (Bailey et al, 1992), 0.05 mol/L Na-citrate buffer of 5.3 pH was used, with sodium hydrogen phosphate as the sodium source for preparing the solution. When this buffer was used to prepare the xylose calibration curve, it was observed that a precipitate was formed after the solution was cooled down, resulting in a blurred sample and absorbance measurement which was too difficult to perform. Therefore, the origin of sodium was changed in the continuation of the research. Instead of sodium hydrogen phosphate, sodium citrate was used in the preparation of the citrate buffer (0.05 mol/L, pH 5.3) and consequently, no precipitation occurred for experiments performed with sodium citrate-based buffer.
3.4. Activity assay
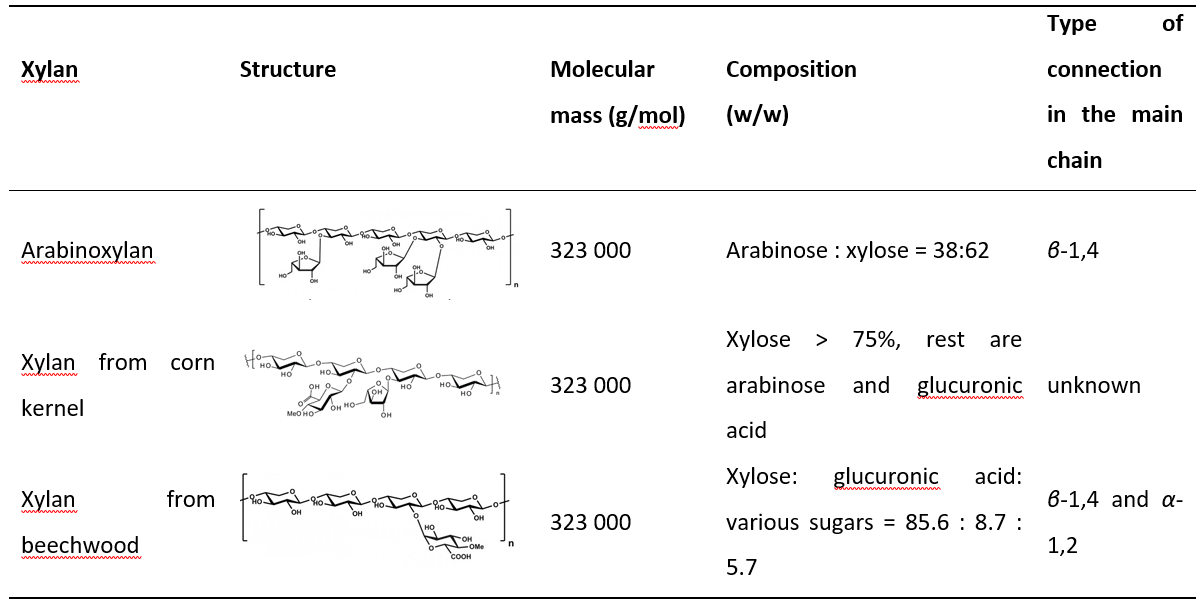
Once the DNS reagent, substrate, and buffer were selected, the activity assay was performed. First, it was necessary to create a calibration curve under new conditions (Figure 1). Correlation of determination of R2 = 0.9934 was calculated for a 95% confidence interval.
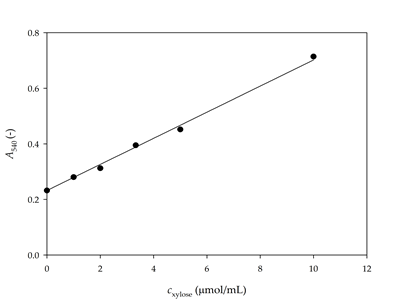
Figure 1. Calibration curve for the determination of xylose concentration with DNS I reagent
To test the method and the chosen conditions, the activity of the xylanase from different sources was determined using 1% arabinoxylan as substrate. This substrate was chosen because, according to the manufacturer, the activities indicated on the enzyme packaging were determined with the same substrate. According to McCleary and McGeough (2015), to compare enzyme activities same substrate, equal concentrations and the same assay procedure should be used. A total of 5 xylanases were tested, two from A. niger, and one from Bacillus SMB, T. longibrachiatum, and T. reesei, respectively. Specific xylanase activities were calculated as the ratio of sugar concentration (determined according to the calibration curve) to incubation time and protein concentration. The obtained results are shown in Table 5. As can be seen, the values obtained were in accordance with the values specified by the manufacturer, which shows that the assay was accurate.
Table 5. Comparison of the activities indicated by the manufacturer and activities measured by the developed assay
As mentioned, enzyme activity is a function of incubation time, sugar concentration, and protein concentration. In the original Bailey method (Bailey et al., 1992), the incubation time was fixed at 5 min. Miller (1959) increased this time to 15 min, suggesting that 5 min is not sufficient for complete colour development. However, both methods assume that the xylose concentration change is linear during incubation. To verify this assumption, a new set of experiments was performed.
3.5. Influence of reaction time on the determination of enzyme activity
The dynamic change in xylose concentration was monitored to verify if 5 min of reaction time was sufficient for the hydrolysis of the substrate. Both selected substrates (arabinoxylan, and xylan from beech wood) were tested. The reactions were performed for 5, 10, 20, 60, 120 and 360 s, increasing the concentration of xylose, as shown in Figure 2.
Figure 2. Dynamic change of xylan consumption and xylose formation for a) 1% arabinoxylan, b) 1% xylan from beechwood and c) 0.01% xylan from beechwood as substrates (● experimental data, ▬ mathematical model of xylane consumption, ▬▬ mathematical model of xylose formation)
When the reaction was carried out using 1% arabinoxylan as a substrate, the concentration of the produced xylose did not increase after 120 s of the reaction. Consequently, the reaction time of the assay should be shortened from 300 s to 120 s in order to obtain more accurate results.
When xylan from beechwood is used, the obtained results show that the xylose concentration remains practically constant after 100 s, which is an additional indication that the reaction time of the assay should be shortened from 300 s to about 100 s when xylan from beechwood is used.
Clearly, incubation time should be shortened from 5 min to 1.5 min to obtain accurate results in the assay.
3.6. Estimation of kinetic parameters
To evaluate the kinetic parameters in the degradation of arabinoxylan and xylan from beechwood, the following solutions of both substrates were prepared (w/w): 0.01%, 0.05%, 0.1%, 0.5% and 1%. The dependence of the initial reaction rate on the xylan concentration was determined using the initial rate method, and inhibition was considered negligible. For both substrates, the reaction kinetics was described by the Michaelis-Menten kinetic model. The estimated parameters of the Michaelis-Menten kinetic model are listed in Table 6.
Table 6. Kinetic parameters of the Michaelis-Menten kinetic model for enzymatic degradation of arabinoxylan and xylan from beechwood
| Parameter | Arabinoxylan | Xylan from beechwood |
| Vmax (U/mL) | 13.46±0.25 | 28.57±2.42 |
| Km (nmol/mL) | 0.70±0.04 | 12.06±1.85 |
The maximum activity of the xylanase for the reaction where xylan from beechwood was used as a substrate is approximately twice as high as that for arabinoxylan. The reason for this is probably the different structure and composition of xylan from different sources. Arabinoxylan is highly branched and consists of xylose and arabinose (62:38). Xylan from beechwood contains 85.6% xylose and the rest are glucuronic acid and sugars. Based on this, it can be concluded that purified xylane from beechwood is a suitable substrate for xylanase assay. The same results were confirmed by McCleary and McGeough (2015).
In addition to estimating the kinetic parameters, a mathematical model of the process was established to describe the change in concentration of xylan and xylose for the reaction performed in a batch reactor (Equations 2 and 3):
where r is the rate of the enzymatic reaction.
The validation of the model was performed using a 1% arabinoxylan solution, and 0.01% and 1% xylan solution from beechwood. To solve the mathematical model of the process, initial concentration of xylose in xylan had to be determined. For this purpose, chemical hydrolysis of the two substrates was performed. After hydrolysis, it was determined that a 1% arabinoxylan solution contained 1.31±0.01 µmol/mL of xylose, a 1% xylan from beechwood solution contained 4.39 µmol/mL of xylose, while a 0.01% xylan from beechwood solution contained 0.044±0.001 µmol/mL of xylose. Simulation results are presented in Figure 2.
A good agreement between experimental results and the model simulation results was obtained, demonstrating the applicability of the proposed model for further development and optimization of the assay.
3.7. Calculation of the xylanase activity
Xylanase activity is usually calculated according to the equation that considers the concentration of xylose formed and the duration of the xylan biotransformation reaction in which xylose is formed (Equation 1). On the other hand, the amount of substrate and enzyme in the sample and the substrate origin play an important role in determining the enzymatic activity (McCleary and McGeough, 2015). To include this in calculation of xylanase activity, an extension of that initial equation (Equation 1) is proposed in Equations 4 and 5.
where c is the enzyme concentration, VT is the total volume of reaction mixture, t is the incubation time, VE is the volume of added enzyme solution, ΔA is the absorbance calculated as the difference between absorbances of the main probe and substrate blank or enzyme blank, d is the diameter of the cuvette (cm), ε is the molar absorption (extinction) coefficient, and DF is the dilution factor.
By measuring the change in absorbance of samples of different concentrations of xylose, the extinction coefficient was determined experimentally to be ε = 0.0624±0.0009 L/(mmol · cm).
By calculating the volume activity of the xylanase using Equations 4 and 5, volume activities obtained were on average 1.2 (Equation 4) and 1.1 (Equation 5) fold higher, respectively, than the volume activity calculated by the original equation used in this study to calculate all previous volume activities.
Because Equation 5 takes into account most of the factors that are critical for determining activity, it was concluded that its use should be most consistent with the used assay to measure the xylanase activity.
To validate the proposed method, the xylanase activity was measured with the crude xylanase extract, obtained after solid-state fermentation, and its activity was determined using proposed equations (Equation 1, Equation 4, Equation 5 and Equation 6) and the developed assay. The following xylanase activities were estimated: Equation 1, V.A. = 14.70±1.41 U/mL, Equation 4, V.A. = 17.79±1.20 U/mL, Equation 5, V.A. = 16.83±1.13 U/mL and Equation 6, V.A. = 16.23±2.29 U/mL.
3.9. The protocol of the newly developed assay for xylanase activity measurement
Based on all obtained results the following assay for measurement of xylanase activity was proposed:
DNS reagent I preparation (according to Gusakov et al., 2011). Prepare 0.02 g/L 3,5-dinitrosalicylic acid in 0.5 mol/L sodium hydroxide solution at 70 °C. After the 3,5-dinitrosalicylic acid is completely dissolved, add 60 g of sodium potassium tartrate (Rochelle salt) gradually until it is completely dissolved. The prepared reagent should be stored in a dark reagent bottle until use.
Buffer (0.05 mol/L Na-citrate buffer pH 5.3) preparation. Dissolve 963 mg of sodium citrate dihydrate and 332 mg of citric acid in 80 mL of distilled water. Mix the content on a magnetic stirrer until the components are completely dissolved. Adjust pH to 5.3 by the addition of 1 mol/L NaOH solution or 1 mol/L HCl solution. Adjust volume to 100 mL with a distilled water solution.
Substrate preparation. Add 1 g of xylan from beechwood to 80 mL of 0.05 mol/L Na-citrate buffer of the 5.3 pH value and dissolve it by stirring at 60 °C. After cooling to 25 °C, add Na-citrate buffer to 100 mL and store prepared solutions at 25 °C for no more than one week.
Enzyme preparation. Dissolve 1 mg of enzyme in 1 mL 0.05 mol/L Na-citrate buffer of the pH value of 5.3. Depending on enzyme activity, enzyme should be diluted if the reaction is too fast to monitor (high activity), or more enzyme should be dissolved if the reaction is to slow (low activity).
Assay procedure (modified Bailey method (Bailey et al., 1992). 0.9 mL of substrate solution and 0.1 mL of enzyme solution should be mixed and heated at 50 °C for 100 s. Add 1.5 mL of the DNS reagent to the mixture. Heat the new mixture at 100 °C for 5 min. Cool the mixture to 25 °C with a water bath and measure the absorbance at λ = 540 nm.
To monitor the spontaneous decomposition of the substrate, a substrate blank should be prepared by using 0.1 mL of 0.05 mol/L Na-citrate buffer pH 5.3 instead of 0.1 mL of enzyme according to the same procedure.
If the dilution is rather small and/or if the sample contains a high level of reducing sugars, an enzyme blank is required (i.e. in samples after fermentation or lignocellulose degradation) instead of a substrate blank. Prepare enzyme blank by heating 0.9 mL of substrate solution to 50 °C for 100 s and then add 1.5 mL of the DNS reagent followed by 0.1 mL of enzyme solution. Heat the mixture to 100 °C for 5 min, cool to 25 °C, and measure absorbance at λ = 540 nm.
Activity calculation (Equation 6):
where Δ A is the absorbance calculated as the difference between absorbances of the main probe and substrate blank or enzyme blank, t is the incubation time (2 min), d is the diameter of the cuvette (cm), ε is the molar absorption (extinction) coefficient ( ε = 0.0624 L/(mmol · cm)), VR is the volume of reaction mixture (2.5 mL), VE is the volume of added enzyme solution (0.1 mL) and DF is the dilution factor.
4. Conclusions
The drawbacks of the 3,5-dinitrosalicylic acid (DNS) assay have been critically analysed to improve the accuracy of the method. Basic DNS assay factors like buffer preparation, substrate origin and concentration, incubation time and reagent preparation were re-evaluated and improved. As a result, a new assay for the measurement of xylanase activity was proposed. Compared to the assays proposed in the literature, the current assay is shorter, does not require the preparation of a calibration curve for xylose, takes into account the concentration of substrate and enzyme in the sample, and provides the equation that accounts for most of the factors that are critical for determining enzyme activity.
Author Contributions:
Marko Božinović: conceptualization, data curation, formal analysis, investigation, writing—review and editing, Tea Sokač: conceptualization, data curation, formal analysis, investigation, methodology, writing—review and editing, Anita Šalić: conceptualization, data curation, methodology, software, visualization, writing—original draft preparation, writing—review and editing, Ana-Marija Dukarić: data curation, investigation, Marina Tišma: supervision, writing—review and editing, Mirela Planinić: funding acquisition, project administration, resources, writing—review and editing, Bruno Zelić: supervision, writing—review and editing
Funding: This work was supported by the European Regional development Fund (ERDF) (grant KK.01.1.1.04.0107).
Conflicts of Interest: The authors declare no conflict of interest
