INTRODUCTION
Pancreatic cancer is a lethal malignancy causing a significant number of human deaths annually (1). The global burden of pancreatic cancer is increasing at an alarming rate and has more than doubled over the past two decades (2). With a poor 5-year survival rate, novel strategies need to be developed for the management of pancreatic cancer (3). Treatment options for pancreatic cancer include surgery, chemotherapy, radiation therapy, and emerging targeted therapies (4). Surgical resection offers the best chance for cure but is feasible for only a minority of patients. Chemotherapy, including regimens like FOLFIRINOX and gemcitabine/nab-paclitaxel, extends survival but is rarely curative (5). Radiation therapy complements other treatments, while targeted therapies like poly (ADP-ribose) polymerase (PARP) inhibitors and immunotherapy show promise in clinical trials (6). Continuous research efforts are necessary to advance early detection, discover new therapeutic targets, and innovate treatment strategies, offering hope to patients affected by this devastating disease (7, 8).
In recent years, non-coding RNAs (ncRNAs) have garnered significant attention in cancer research, with a particular focus on their role in tumorigenesis, including pancreatic cancer (9). Among ncRNAs, long non-coding RNAs (lncRNAs), a subclass characterized by their extended nucleotide sequences exceeding 200 bases, have emerged as key regulators of fundamental cancer hallmarks (10). It is well-established that lncRNAs exert diverse control over various biological processes by modulating the expression of target genes at transcriptional, post-transcriptional/translational, or epigenetic levels (11). Importantly, dysregulated lncRNA expression has been closely linked to the initiation and progression of tumors (12), as lncRNAs play pivotal roles in modulating critical aspects of cancer cell behavior, such as proliferation, apoptosis, migration, invasion, and microenvironment remodeling (13, 14). Additionally, lncRNAs contribute to the maintenance of stemness in various human cancers (15). Given the central role of lncRNAs in cancer regulation, accumulating evidence suggests that lncRNA-based cancer therapeutics may soon transition from a theoretical concept to a tangible reality.
TINCR, an acronym for Terminal differentiation-induced non-coding RNA, is a type of RNA molecule that does not code for proteins. It is located on chromosome 19p13.3 and has been identified as having a significant role in several cancer types, such as breast cancer, lung cancer, gastric cancer, and liver cancer. This non-coding RNA's involvement in various cancers highlights its importance in understanding and potentially targeting cancer-related processes (16, 17). Notably, TINCR demonstrates varying effects on the progression and development of different types of human cancers, highlighting its context-dependent characteristics (18). Despite these findings, our understanding of the precise impact of TINCR on the biological characteristics of pancreatic cancer cells remains limited. This knowledge gap emphasizes the importance of conducting a comprehensive investigation into TINCR's role within the context of pancreatic cancer. In accordance with these observations, our study is dedicated to a thorough exploration of the involvement of the long non-coding RNA TINCR in controlling the growth and metastasis of human pancreatic cancer. Our research initiative seeks to unravel the intricate mechanisms through which TINCR influences the pathophysiology of pancreatic cancer.
EXPERIMENTAL
Cell lines
Pancreatic cancer cell lines like PaCa-2 (RRID:CVCL_0428), PL45 (RRID:CVCL_3567), PANC-1 (RRID:CVCL_0480), and BxPC-3 (RRID:CVCL_0186) were obtained from the American Type Culture Collection USA. The human normal pancreatic ductal epithelial cell line (H6C7) was obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (China). Cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10 % fetal bovine serum (FBS) and streptomycin-penicillin (100 U mL–1). The cells were maintained in a humidified CO2 incubator at 37 °C with 5 % CO2.
Chemicals and kits
Dulbecco’s modified Eagle’s medium was purchased from Gibco, Gaithersburg, USA). Lipofectamine 2000 was purchased from Invitrogen (USA.
SYBR PCR master mix was purchased from Takara, Japan. PcDNA3.1 overexpression vector was purchased from Addgene, USA. Trizol reagent, Revert Aid First Strand cDNA synthesis kit, Cell Counting Kit-8 assay kit, EdU solution, Annexin V-FITC Apoptosis Detection Kit, propidium iodide, medetomidine, midazolam, and butorphanol were all obtained from Sigma Aldrich, USA.
Transfection
The overexpression vector for TINCR was constructed using pcDNA3.1. TINCR (NM_001396408.1) was amplified by PCR from cDNA extracted from PANC-1 cells, employing specific primers: TINCR forward primer 5'-CGGATCCAGAGCTGGAGCCGGAGCCGGGC-3' (with BamHI restriction site) and TINCR reverse primer 5'-TGAATTCTAGTTCCAAGCTGGGTGATCC-3' (with an EcoRI restriction site). PCR was performed in Veriti 96 well thermocycler (ThermoFisher Scientific, USA), with program conditions: 95 °C for 5 min, 30 cycles of (denaturation: 95°C for 30 sec, annealing: 63°C for 60 sec, and extension: 72°C for 1 min), final extension: 72 °C for 10 min, and hold temperature at 4 °C. The resulting PCR product was separated on a 1.2 % agarose gel and purified using a Gel Purification Kit (Qiagen, USA). Subsequently, the purified product underwent digestion with BamHI and EcoRI restriction enzymes and was again purified using the Gel Purification Kit. The doubly digested amplicon was then ligated with the BamHI/EcoRI-digested pcDNA3.1 vector using T4 DNA ligases, resulting in the generation of the pcDNA-TINCR construct. The transfection procedure was carried out in a serum-free medium using Lipofectamine 2000, following the manufacturer's recommended protocol. Approximately 6 hours after the initial transfection, the cells were replenished with a fresh complete medium. Following this medium change, the cells were allowed to incubate for approximately 48 hours. After this incubation period, the cells were harvested and collected for subsequent analysis. To serve as a control group for comparison, cells transfected with the pcDNA3.1 vector alone (without the TINCR insert) were included. These cells, designated as the negative control cells, provided a reference point for evaluating the effects of TINCR overexpression in the experimental group.
Expression analysis via qRT-PCR
To analyze the expression of the target gene, qRT-PCR was employed. Firstly, total RNA was extracted from the cell samples using Trizol reagent, following the manufacturer's guidelines for optimal RNA isolation. Isolated RNA (100 ng) was reversely transcribed using the Revert Aid First Strand cDNA synthesis kit according to the manufacturer’s instructions. For the quantification of gene expression, a qRT-PCR approach was adopted. In this method, the SYBR PCR master mix was used in conjunction with the QuantStudio 3.0 PCR system from Applied Biosystems. In order to standardize and normalize the data, the GAPDH gene was utilized as an internal control in the expression study. The qRT-PCR primers used were: e-cadherin (forward 5'-GTCAGTTCAGACTCCAGCCC-3' and reverse 5'-AAATTCACTCTGCCCAGGACG-3'); α-catenin (forward 5'-CAACCCTTGTAAACACCAAT-3' and reverse 5'-CCTTCTCCAAGAAATTCTCA-3'); vimentin (forward 5'-TCTACGAGGAGGAGATGCGG-3' and reverse 5'-GGTCAAGACGTGCCAGAGAC-3'); fibronectin (forward 5'-CCCACCGTCTCAACATGCTTAG-3' and reverse 5'-CTCGGCTTCCTCCATAACAAGTAC-3'); GAPDH (forward 5'-ACCACAGTCCATGCCATCAC-3' and reverse: 5'-TCCACCCTGTTGCTGTA-3'). The reaction conditions were: 94 °C for 10 minutes; 35 cycles of 94 °C for 15 s, 55 °C for 40 seconds, and 72 °C for 20 seconds. The relative expression levels of TINCR were calculated using the widely accepted 2-ΔΔCt method (19).
CCK-8 assay
To evaluate the viability of PANC-1 cells following transfection with either the pcDNA- TINCR construct or the control vector (pcDNA3.1), a Cell Counting Kit-8 (CCK-8) assay was employed. Initially, a total of 105 PANC-1 cells were seeded into each well of a 96-well plate. These cells were then allowed to incubate for a 24-hour period at a temperature of 37 °C to facilitate adherence and growth. Subsequently, the CCK-8 solution was introduced into each well, and the incubation continued at 37 °C for an additional hour. This solution is known to contain a colorimetric reagent that becomes metabolically active in living cells. Following the incubation period, the optical density at 570 nm (OD570) was measured for each well using a microplate reader (ELx808™, Agilent, USA). This OD570 reading served as an indicator of cell viability. Essentially, a higher OD570 value indicates a greater number of viable cells, while a lower value suggests reduced cell viability.
EdU assay
To delve into the impact of TINCR overexpression on the proliferation of PANC-1 cancer cells, we employed an EdU Apollo in vitro flow cytometry kit in line with the manufacturer's recommended procedures. Initially, the transfected cells (2 × 103 cells/well) were cultured in a 12-well plate and allowed to incubate for a 24-hour period at 37 °C. This incubation period was essential for facilitating cell adherence and establishing the experimental conditions. Subsequently, the cells were exposed to an EdU (5-Ethynyl-2'-deoxyuridine) solution at a concentration of 50 µmol L–1. EdU is a thymidine analog that is incorporated into the DNA of actively dividing cells during DNA synthesis. The cells were allowed to incubate with the EdU solution for 2 hours. Following the incubation with EdU, the cells underwent a series of processing steps. They were first washed with phosphate-buffered saline (PBS) and then fixed using a 4 % paraformaldehyde solution. To visualize the EdU-labeled cells, they were stained using Apollo staining solution from San Francisco, CA, for a duration of 25 minutes. Additionally, the cells were counterstained with DAPI (4',6-diamidino-2-phenylindole) to label the cell nuclei. The final step involved visualizing the cells under a fluorescence microscope (Evos FL, ThermoFisher Scientific, USA) to examine the nuclear fluorescence. This allowed for the identification and quantification of cells that had incorporated EdU, indicating active DNA synthesis and, by extension, cell proliferation.
Annexin V/PI staining assay
To assess the impact of TINCR overexpression on apoptosis in PANC-1 cancer cells, a series of experiments were conducted, specifically utilizing an Annexin V/PI staining assay. This assay was employed to distinguish between early and late apoptotic cells, as well as non-apoptotic cells. Initially, PANC-1 cells (2 × 103 cells/well) were transfected with pcDNA- TINCR, and the negative control cells were cultured in DMEM for a 24-hour period within a 12-well plate, maintaining a temperature of 37 °C. Following the incubation period, the cells were harvested and subjected to a thorough washing regimen, involving three washes with phosphate-buffered saline (PBS) to ensure the removal of extraneous cellular debris. The Annexin V-FITC/PI double staining procedure was carried out according to the manufacturer's instructions, employing the Annexin V-FITC Apoptosis Detection Kit. Subsequently, the stained cells were analyzed using BD FACSCanto™ II Flow Cytometry System, BD (USA). Flow cytometry data were processed and analyzed using FlowJo version 6.0 software (BD), which facilitated the quantification of the relative percentage of apoptotic cells within the experimental and control groups.
Wound healing assay
To investigate the migratory behavior of PANC-1 cancer cells overexpressing TINCR in comparison to control cells, a wound-healing assay was conducted. This assay allowed for the evaluation of cell migration by tracking the closure of a scratched "wound" over time. Initially, 5 × 105 PANC-1 cells transfected with pcDNA- TINCR or the control vector were seeded into each well of a six-well plate. Following cell seeding, a 24-hour incubation period at 37 °C was allowed to promote cell adherence and growth. Subsequently, the serum-free medium was carefully aspirated to create a consistent and linear "wound" on the cell monolayer. This was achieved by gently scratching the cell surface with a sterile pipette tip. The resulting scratch, representing the wound, was then immediately documented using a light microscope, with images captured for reference. The plated cells were then incubated at 37 °C for an additional 24 hours. During this incubation period, cell migration and proliferation would contribute to the gradual closure of the wound. After the incubation, the wound area was re-evaluated and photographed under the microscope (CKX53, Olympus, USA). By comparing the initial and final images of the scratch, the extent of wound closure could be quantified. This provided valuable information about the migratory capacity of PANC-1 cells in response to TINCR overexpression, as evidenced by the degree of wound closure.
Transwell assay
We conducted a Transwell assay to investigate the invasive potential of transfected PANC-1 cancer cells. In this assay, we utilized Matrigel-coated Transwell chambers with 8 µm pores. Approximately 2.5 × 104 transfected cells were seeded in the upper chamber, containing serum-free medium, while the lower chamber contained only serum-free medium. Following 24-hour incubation at 37 °C, cells that had successfully invaded through the Matrigel and reached the lower chamber were fixed with 70 % ethanol. Subsequently, these cells were stained with 0.2 % crystal violet and examined under a microscope (CKX53, Olympus, USA) to quantify their invasiveness. This assay provided crucial insights into the ability of these cells to invade and penetrate barriers resembling the extracellular matrix, a critical aspect of cancer cell behavior.
Western blotting
To extract proteins from the transfected PANC-1 cancer cells, we employed the RIPA lysis and extraction buffer, obtained from Thermo Fisher Scientific. The protein content of each extract was quantified using the Bradford assay. Next, 35 µg of protein samples were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The resolved proteins were subsequently transferred onto activated polyvinylidene difluoride (PVDF) membranes. To ensure specific antibody binding and minimize non-specific interactions, the PVDF membranes were blocked using 5 % skimmed milk powder for a duration of 1 hour. Following blocking, the PVDF membranes were incubated overnight at 4 °C with primary antibodies: anti-Bcl-2 antibody (Sigma Aldrich, SAB4500003, 1:1000 dilution, USA) or anti-Bax antibody (Sigma Aldrich, SAB4502548, 1:1000 dilution) or anti-E-cadherin (CST, 3195, 1:800 dilution), anti-alpha catenin (Sigma Aldrich, C2081, 1:1000 dilution), anti-vimentin (Abcam, ab137321, 1:700, USA), anti-fibronectin (Sigma Aldrich, F3648, 1:1000), anti-β-catenin (Abcam, ab6302, 1:1000), anti-p-GSK3β (CST, 9336, 1:1000) and anti-GAPDH (Sigma Aldrich, G9545, 1:1000) was used as a housekeeping control. Following the washing step, horse radish peroxidase (HRP) conjugated anti-rabbit IgG secondary antibodies (CST, 7074 1:4000 dilution, USA) was used and it was detected using the ECL chemiluminescence method. Finally, protein bands were visualized using UVP ChemStudio Western Blot Imaging Systems (Analytik Jena, USA) and subsequently analyzed using ImageJ software (version 1.48u; National Institutes of Health, USA). GAPDH was employed as an endogenous control to normalize protein expression levels.
In vivo xenograft growth assay in nude mice
All animal experiments were conducted in strict accordance with institutional guidelines for the ethical care and use of laboratory animals. The experimental protocol was duly reviewed and approved by the Liaoning Cancer Hospital & Institute (Approval Number: LCH/31/2019-23).
The mice were housed in the Cancer Hospital of China Medical University, maintaining specific pathogen-free conditions. Environmental parameters included a controlled temperature of 23 ± 2 °C, relative humidity of 55 ± 10 %, and a 12-hour light/12-hour dark cycle. Food and water were provided ad libitum, and the mice's health was monitored at least once daily after tumor injection.
Male BALB/c nude mice, aged 4–6 weeks, were randomly separated into two groups, each comprising 8 animals:
Group I: Nude mice were subcutaneously injected with 2 × 106 transfected cells (PANC-1 empty vector) on one side of the posterior flank.
Group II: Mice were subcutaneously injected with 2 × 106 transfected cells ( TINCR-PANC-1) on one side of the posterior flank.
At the end of the experimental period, on day 25, all surgical procedures were performed under anesthesia induced by medetomidine (0.3 mg kg–1), midazolam (4.0 mg kg–1), and butorphanol (5.0 mg kg–1). Euthanasia of mice was conducted using CO2 inhalation at the study's conclusion. Tumor volumes were assessed by measuring two perpendicular diameters with calipers and calculated using the formula V = ( a × b)/2, where a represents the small diameter and b the large diameter. Finally, xenograft tumors were homogenized to create a uniform mixture, and then centrifuged to separate solid cellular components from the liquid supernatant. This supernatant was subsequently employed for Western blot analysis to gauge specific protein expression levels.
Statistical analysis
The obtained results were presented as mean values with positive standard deviation error bars (mean + SD). To ascertain the statistical significance of our findings, we conducted rigorous statistical analyses utilizing GraphPad Prism 7.0 (USA). Unpaired Student's t-tests determined differences between two experimental cell groups. For comparisons involving three or more groups, a one-way ANOVA followed by Tukey's post hoc test was applied. In our presentation of the results, significance levels were indicated as follows: * p < 0.05 for statistically significant results and ** p < 0.01 for highly significant findings.
RESULTS AND DISCUSSION
Downregulation of TINCR expression in PANC-1 cells
LncRNAs have emerged as intriguing regulators of various biological processes, despite their inability to code for proteins (20). Accumulating evidence has underscored their impact on key aspects of human health and disease, with cancer being a prominent area of investigation (21). Dysregulation of lncRNAs has been implicated in the initiation and progression of multiple pathological conditions, including various cancers. Particularly in the context of cancer, lncRNAs have been identified as influential players, modulating critical cellular behaviors such as proliferation, invasion, and metastasis (22).
In our study, we delved into the role of the long non-coding RNA TINCR in the context of pancreatic cancer. Notably, previous research has linked TINCR to cancer suppression in other malignancies, such as melanoma and breast cancer, shedding light on its potential as a tumor suppressor (23, 24).
We aimed to investigate the expression levels of TINCR in various pancreatic cancer cell lines, including PaCa-2, PL45, PANC-1, and BxPC-3, in comparison to H6C7, a human normal pancreatic ductal epithelial cell line. This was accomplished through qRT-PCR analysis.
The qRT-PCR results yielded compelling findings, indicating a noteworthy downregulation of TINCR expression in all the examined pancreatic cancer cell lines when compared to H6C7 (Fig. 1a). This downregulation was found to be statistically significant ( p = 0.04), underscoring the potential importance of TINCR in the context of pancreatic cancer progression. Remarkably, among the tested pancreatic cancer cell lines, PANC-1 exhibited the lowest levels of TINCR transcript expression. This finding led us to select PANC-1 cells for further experimentation due to their pronounced downregulation of TINCR and its potential implications in the context of pancreatic cancer biology.
In summary, the qRT-PCR analysis unveiled a consistent downregulation of TINCR expression in PANC-1 cells, highlighting its potential relevance as a key regulatory molecule in pancreatic cancer and underscoring its candidacy for further investigations in subsequent experiments.
Suppression of pancreatic cancer cell proliferation and viability by TINCR overexpression
TINCR exhibits aberrant expression patterns across various tumor types, suggesting a pivotal role in influencing either oncogenic or tumor-suppressive pathways, thereby influencing tumor initiation and progression. In-depth investigations have unveiled distinct associations between TINCR and breast cancer, where it emerges as an oncogenic player (25). On the other hand, TINCR exerts inhibitory effects on both proliferation and invasion in lung cancer by modulating the miR-544a/FBXW7 axis (26). Here, we endeavored to investigate the influence of TINCR on the proliferation and viability of PANC-1 cells.
To investigate the impact of TINCR on the proliferation and viability of PANC-1 cells, we initiated TINCR overexpression in PANC-1 cells, and the success of this overexpression was confirmed through qRT-PCR analysis (Fig. 1b). Subsequently, we sought to determine whether the upregulation of TINCR had any influence on the viability and proliferation of PANC-1 cells. To accomplish this, we conducted a Cell Counting Kit-8 (CCK-8) assay, a widely used method for assessing cell viability. The results of the CCK-8 assay revealed a noteworthy and statistically significant decrease in the viability of PANC-1 cancer cells overexpressing TINCR in comparison to their corresponding negative control cells (Fig. 1c). This decline in viability strongly suggests that TINCR exerts a suppressive effect on the growth of PANC-1 cells.
Furthermore, we examined the incorporation of EdU (5-Ethynyl-2'-deoxyuridine), a thymidine analog, into PANC-1 cancer cells, as it serves as a marker for actively proliferating cells. Importantly, the overexpression of TINCR was found to substantially reduce the incorporation of EdU into PANC-1 cancer cells (Fig. 1d). This decline in EdU incorporation provides additional evidence of diminished cell proliferation associated with TINCR overexpression.
In summary, our findings suggest that TINCR plays a pivotal role in inhibiting the proliferation and viability of PANC-1 cells. These results shed light on the potential therapeutic relevance of TINCR in pancreatic cancer management.
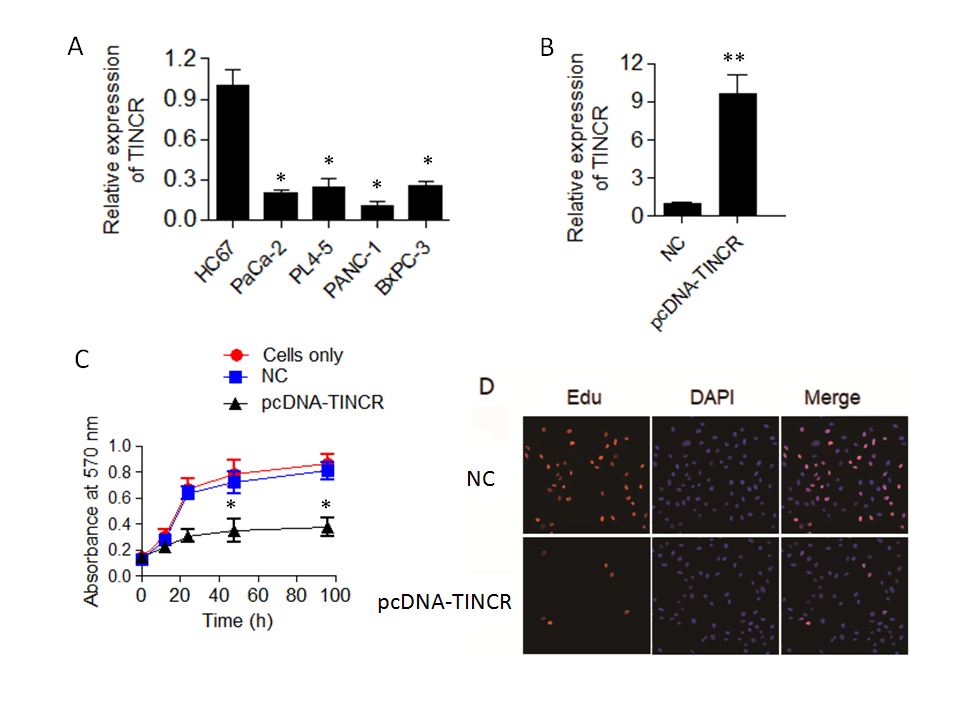
Fig. 1. TINCR expression and its impact on pancreatic cancer cell proliferation. a) Expression levels of long non-coding RNA TINCR were analyzed in various pancreatic cancer cell lines (PaCa-2, PL45, PANC-1, and BxPC-3) compared to the human normal pancreatic ductal epithelial cell line, H6C7, using qRT-PCR. Results demonstrate significant downregulation of TINCR expression in all pancreatic cancer cell lines; b) TINCR overexpression in PANC-1 cells confirmed by qRT-PCR; c) cell Counting Kit-8 (CCK-8) assay was performed to assess the impact of TINCR overexpression on the proliferation of PANC-1 cancer cells. TINCR overexpression led to a significant reduction in cell proliferation compared to the corresponding negative control cells; d) EdU incorporation assay showed a marked decrease in the incorporation of EdU into PANC-1 cancer cells overexpressing TINCR, indicating a loss of cell viability. * p < 0.05 and ** p < 0.01, compared to control. Data are representative of at least three independent experiments.
TINCR -induced apoptosis in PANC-1 cells
In our quest to unravel the underlying mechanisms responsible for the tumor-suppressive effects of TINCR in PANC-1 cells, we turned our attention to the intricate process of cell apoptosis. To investigate this, we employed the Annexin V-FITC/PI staining method, a robust technique for discerning apoptotic events within the cellular context. The results of our Annexin V-FITC/PI staining revealed a compelling story. The overexpression of TINCR in PANC-1 cells had a profound and statistically significant impact ( p = 0.02) on the induction of apoptosis, with both early and late apoptotic cells displaying a remarkable increase in comparison to control cancer cells (Fig. 2a). This shift towards heightened apoptosis was indicative of TINCR's potent pro-apoptotic role in these cells.
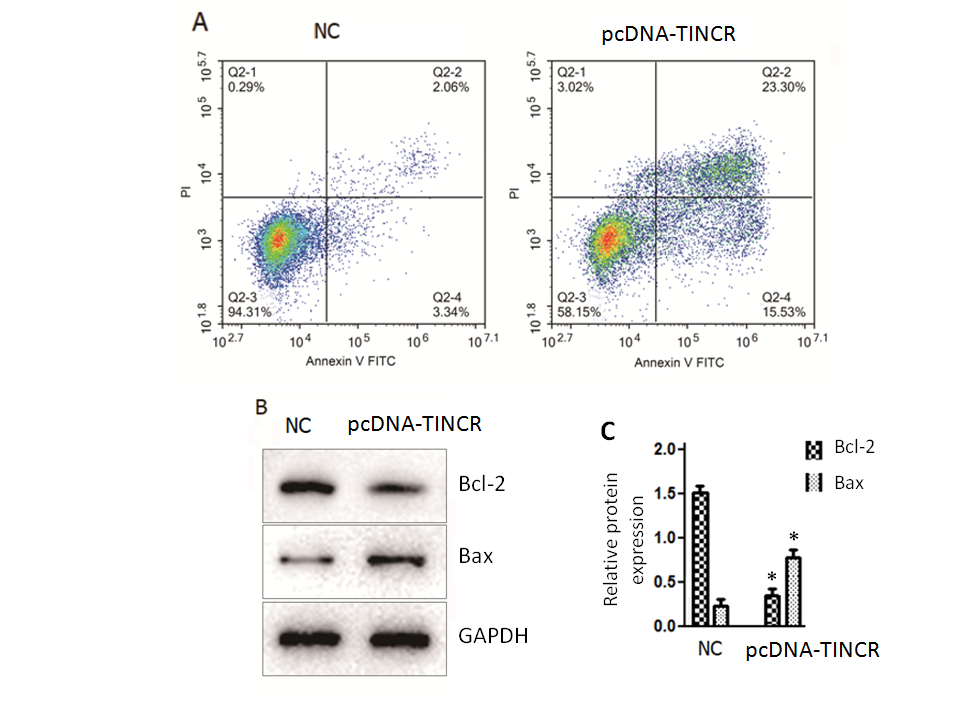
Fig. 2. TINCR induces apoptosis in PANC-1 cells. a) Apoptosis analysis was conducted using Annexin V-FITC/PI staining in PANC-1 cells overexpressing TINCR and control cells. TINCR overexpression increased the percentage of both early and late apoptotic cells compared to the control, with the apoptotic cell percentage rising upon TINCR overexpression compared to control cells; b) Western blot analysis of PANC-1 cancer cells overexpressing TINCR and control cells revealed upregulation of Bax protein expression and marked repression of Bcl-2 protein expression in TINCR-overexpressing cells compared to the negative control cells; c) densitometry analysis of Western blot. * p < 0.05, compared to control. Data represent at least three independent experiments.
Furthermore, a closer examination of the apoptotic cell percentage underscored the significance of TINCR overexpression. Notably, the apoptotic cell percentage exhibited a substantial increase following TINCR overexpression when compared to control conditions, reinforcing the notion that TINCR exerts a pro-apoptotic influence on PANC-1 cancer cells. Complementing these findings, our Western blotting analysis provided molecular insights into the apoptotic process. PANC-1 cancer cells that upregulated TINCR displayed a marked increase ( p < 0.03) in the expression of the pro-apoptotic protein Bax, while the expression of the anti-apoptotic protein Bcl-2 was notably repressed when compared to negative control cells (Fig. 2b). This shift in the balance of pro- and anti-apoptotic proteins further substantiated the role of TINCR in driving apoptotic pathways within PANC-1 cells.
In summary, our comprehensive examination of apoptosis in PANC-1 cells unveiled a pivotal role for TINCR in enhancing apoptosis, as evidenced by the increased percentages of apoptotic cells and the molecular alterations observed in Bax and Bcl-2 expression. These findings shed light on the potential mechanisms through which TINCR exerts its tumor-suppressive effects in pancreatic cancer.
Impact of TINCR on the migration and invasion of PANC-1 cells
Metastasis is a pivotal hallmark of cancer, contributing significantly to disease progression and treatment resistance (27). In our study, we extended our investigation to explore the influence of TINCR on the metastatic potential of PANC-1 cells. Metastasis is intricately linked to cell migration and invasion, both of which are essential for cancer cells to navigate through the body and establish secondary tumors (28, 29). Our results revealed that TINCR overexpression led to a substantial reduction in the rates of metastasis in vitro. Additionally, TINCR upregulation triggered notable changes in the expression of EMT markers, a process often associated with enhanced metastatic capability. These findings suggest that TINCR may act as a regulator of pancreatic cancer metastasis, which is consistent with recent research implicating TINCR in breast cancer metastasis (30).
Our investigation extended to the evaluation of how TINCR influences the migratory and invasive properties of PANC-1 cells in an in vitro setting. To accomplish this, we employed a wound-healing assay to assess cell migration and a Transwell invasion assay to evaluate cell invasion.
In the wound-healing assay, we observed a noteworthy reduction ( p = 0.04) in the migration of PANC-1 cancer cells when TINCR was overexpressed (Fig. 3a). Specifically, the percentage of cell migration was significantly diminished by over 70 % in PANC-1 cells that were overexpressing TINCR compared to the negative control cells. This substantial decrease in cell migration underscored the potent inhibitory effect of TINCR on the migratory capacity of these PANC-1 cells.
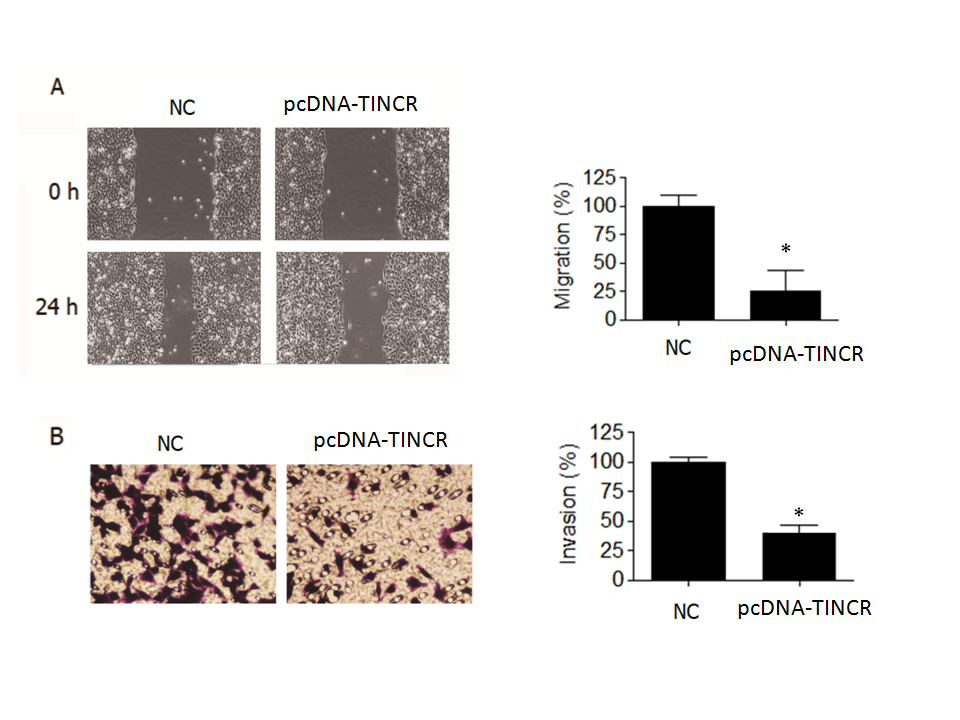
Fig. 3. TINCR inhibits metastasis of PANC-1 cells. a) Wound-healing assay was conducted to assess the effect of TINCR overexpression on the migration of PANC-1 cells. TINCR overexpression significantly reduced cell migration, with over 70 % decrease in cell migration observed in PANC-1 cells overexpressing TINCR compared to negative control cells; b) Invasion assay demonstrated a significant reduction in the invasion capability of PANC-1 cancer cells upon TINCR upregulation. PANC-1 cells overexpressing TINCR exhibited only one-fourth of the invasion rate of negative control cells. * p < 0.05, compared to control. Data are representative of at least three independent experiments.
Furthermore, our investigation into cell invasion yielded equally compelling results. The Transwell invasion assay demonstrated a significant reduction in the invasive potential of PANC-1 cancer cells when TINCR was upregulated (Fig. 3b). More specifically, PANC-1 cells that overexpressed TINCR exhibited an invasion rate that was merely one-fourth that of the negative control cells. This marked decrease in invasive capacity provided robust evidence of TINCR's inhibitory influence on the invasiveness of PANC-1 cells.
In summary, our findings underscore the pivotal role of TINCR in impeding both the migration and invasion of PANC-1 cells. The substantial reductions observed in cell migration and invasion rates upon TINCR upregulation emphasize its potential significance in modulating the aggressive behavior of PANC-1 cells, with potential implications for the suppression of metastasis.
TINCR inhibits EMT in PANC-1 cells
In our exploration of the role of TINCR in pancreatic cancer, we sought to understand its influence on the expression of key markers associated with EMT, a fundamental process in cancer progression. Our investigation involved the assessment of transcript levels of specific EMT markers. The results unveiled a compelling pattern of gene expression changes associated with TINCR.
Firstly, we observed a notable increase in the expression of E-cadherin and α-catenin (Fig. 4a,b). E-cadherin is a critical cell adhesion molecule associated with epithelial characteristics, and α-catenin is a protein involved in adherens junctions, both of which are indicative of an epithelial phenotype. This downregulation suggests that TINCR promotes the retention of epithelial features within PANC-1 cells. Conversely, the expression levels of vimentin and fibronectin exhibited a significant decrease (Fig. 4c,d). Vimentin and fibronectin are markers typically associated with the mesenchymal state, which is characterized by increased motility and invasiveness. The increase in their expression levels suggests that TINCR inhibits the acquisition of mesenchymal traits in these cells.
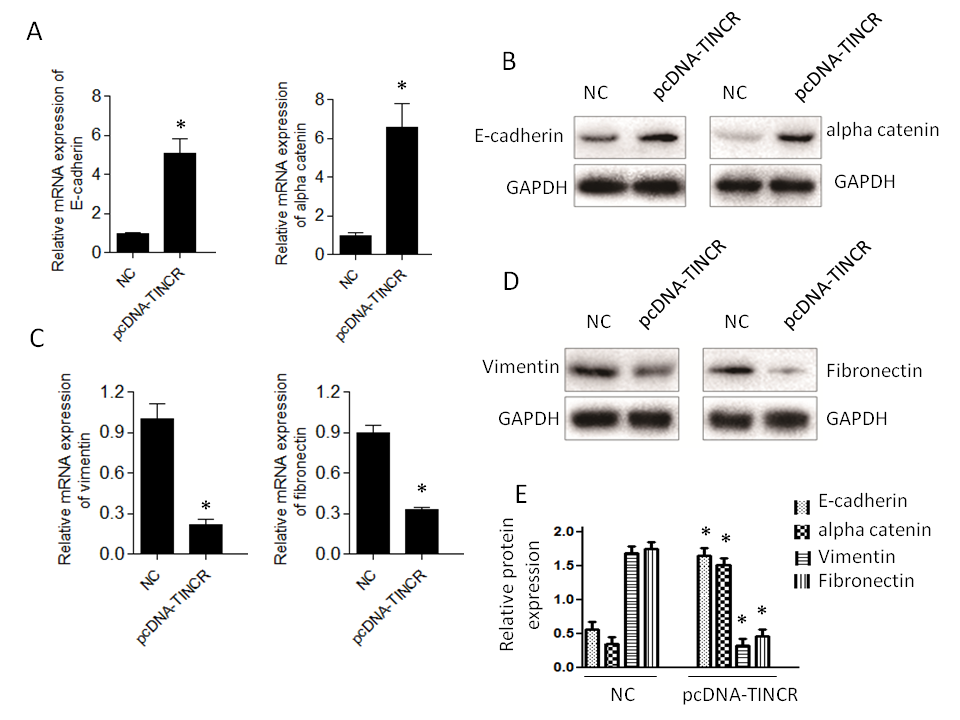
Fig. 4. TINCR modulates epithelial-to-mesenchymal transition (EMT) marker expression. a) and b) qRT-PCR and Western blotting analysis of epithelial (E-cadherin and α-catenin) markers in PANC-1 cells overexpressing TINCR compared to the control cells; c) and d) qRT-PCR and Western blotting analysis of mesenchymal markers (vimentin and fibronectin) in PANC-1 cells overexpressing TINCR compared to the control cells; e) densitometry analysis of Western blot. * p < 0.05, compared to control . Data represent at least three independent experiments.
In summary, our findings indicate that TINCR plays a pivotal role in promoting an epithelial phenotype and restraining the mesenchymal features typically associated with EMT in PANC-1 cells. These observations provide valuable insights into the potential mechanisms through which TINCR influences cancer progression and metastasis in this context.
The inhibition of the Wnt/β-catenin signaling pathway by TINCR>
While our research has confirmed that TINCR can induce EMT in PANC-1 cells, the specific signaling pathways involved have remained unclear. To investigate TINCR 's potential impact on signaling cascades, we focused on the Wnt/β-catenin pathway. The decision to investigate the Wnt/β-catenin pathway in the context of TINCR-induced EMT is underpinned by its known involvement in orchestrating key events during cancer progression (31). Several studies have highlighted the Wnt/β-catenin pathway as a crucial mediator of EMT. Using Western blot experiments, we systematically examined the expression patterns of key proteins in the Wnt/β-catenin pathway, specifically β-catenin and p-GSK3β. Elevated TINCR expression was associated with a notable increase in p-GSK3β levels and a simultaneous decrease in β-catenin protein expression (Fig. 5). In summary, our findings indicate that TINCR can inhibit the Wnt/β-catenin signaling pathway, highlighting its role in inhibiting the EMT process in PANC-1 cells. This contributes significantly to our understanding of the complex molecular dynamics at play in these cells.
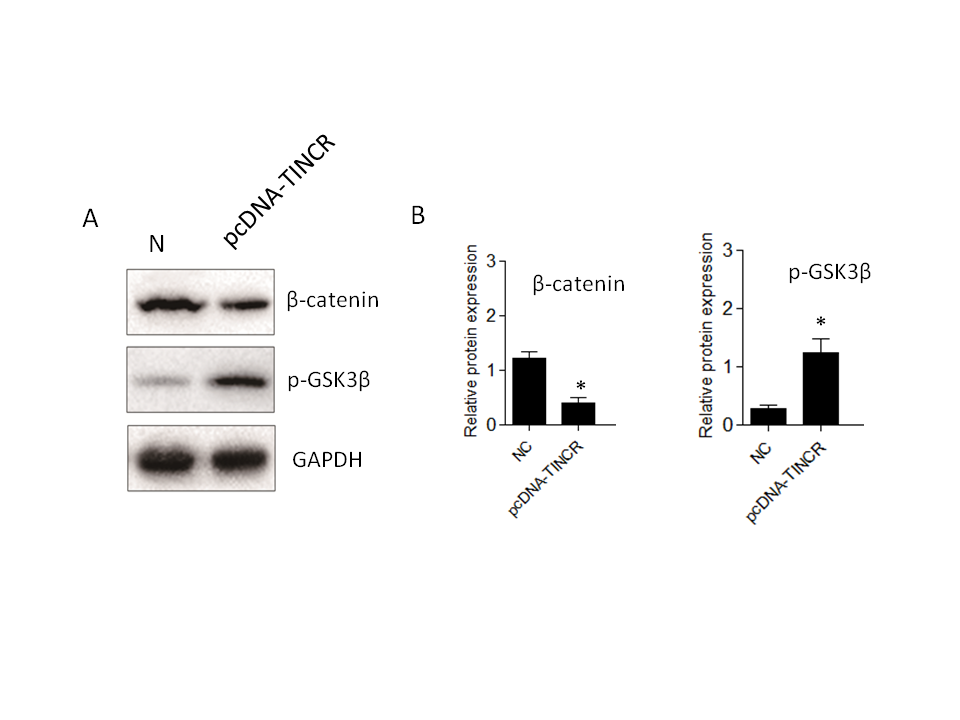
Fig. 5.TINCR-mediated inhibition of the Wnt/β-catenin signaling pathway in PANC-1 cells. a) Western blot analysis of key proteins in the Wnt/β-catenin pathway. Elevated TINCR expression correlated with increased p-GSK3β levels, while simultaneously decreased β-catenin protein expression; b) and c) densitometry analysis of Western blot. * p < 0.05, compared to control . Data represent at least three independent experiments.
TINCR prevents tumor growth, induces apoptosis, and inhibits Wnt/β-catenin in mice
Our study unveils the multifaceted impact of TINCR overexpression in a pancreatic cancer mice model, shedding light on potential therapeutic strategies. TINCR emerges as a potent modulator, impeding tumor growth and reducing tumor size, suggesting its promise as a therapeutic target for pancreatic cancer (Fig. 6). Notably, TINCR demonstrates its ability to promote apoptosis, as evidenced by increased pro-apoptotic protein Bax levels and decreased anti-apoptotic protein Bcl-2 levels, indicating its role in initiating programmed cell death – an essential mechanism to curtail cancer cell survival. Furthermore, our research reveals TINCR's involvement in inhibiting the Wnt/β-Catenin signaling pathway, a key regulator of cell proliferation and cancer progression, presenting a potential avenue for targeted therapies.
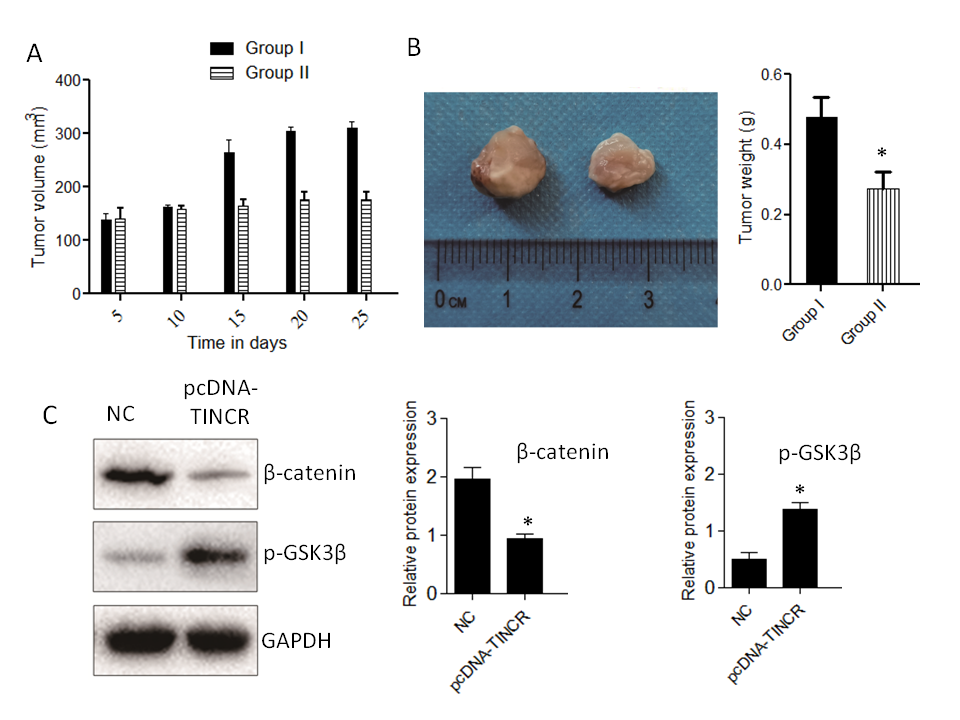
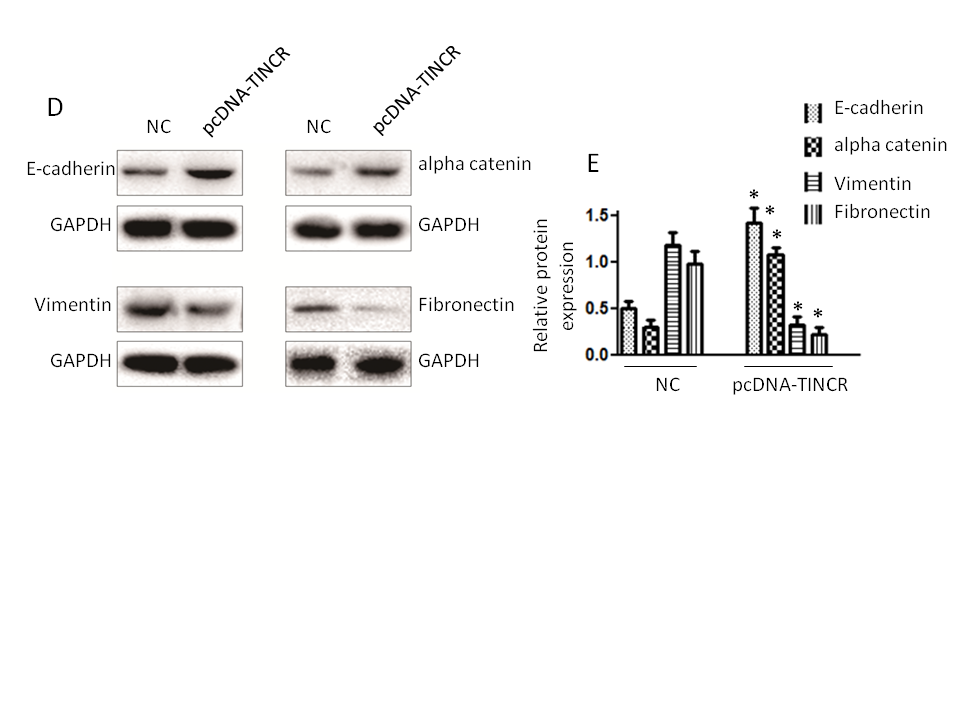
Fig. 6. Impact of TINCR on pancreatic cancer progression in mice. a) Tumor growth inhibition: The effect of TINCR on tumor growth in mice; b) tumor size reduction: The influence of TINCR on tumor size during the study; c) Wnt/β-Catenin inhibition: TINCR's role in modulating the Wnt/β-Catenin signaling pathway in mice; d) inhibition of EMT markers: The impact of TINCR on EMT markers in pancreatic cancer; e) densitometry analysis of Western blot. * p < 0.05, compared to control . Data represent at least three independent experiments.
In our in vivo xenograft growth assay in nude mice, key outcomes include substantial inhibition of tumor growth and consistent reduction in tumor size in the TINCR-overexpressing Panc-1 cell group compared to the control group. Enhanced activation of the Wnt/β-Catenin signaling pathway, as indicated by increased p-GSK3β levels and decreased β-catenin protein expression in the TINCR-overexpressing group, further supports the involvement of TINCR in modulating critical signaling pathways. Additionally, TINCR demonstrates an influence on epithelial-mesenchymal transition (EMT) markers, promoting the preservation of epithelial traits and hindering the acquisition of mesenchymal characteristics in PANC-1 cells.
These findings provide compelling evidence for the therapeutic potential of TINCR in pancreatic cancer. The observed effects on tumor growth, apoptosis, Wnt/β-Catenin signaling, and EMT present a comprehensive understanding of TINCR's impact, paving the way for innovative therapeutic strategies in pancreatic cancer management. Further exploration into TINCR-based interventions holds promise for the development of targeted therapies aimed at improving outcomes for pancreatic cancer patients.
CONCLUSIONS
In summary, our study highlights the noteworthy downregulation of lncRNA TINCR in PANC-1 cells. The overexpression of TINCR emerged as a potent inhibitor of cancer cell proliferation, primarily through the induction of apoptosis. Furthermore, our investigation revealed a significant decrease in the migratory and invasive capabilities of PANC-1 cells following TINCR upregulation. Furthermore, TINCR inhibits tumor growth, triggers apoptosis, and inhibits the Wnt/β-Catenin pathway in mice. These findings pave the way for further research into TINCR's mechanisms and its potential as a targeted therapy for pancreatic cancer. Understanding TINCR's nuanced role could lead to more effective strategies against this challenging malignancy, offering hope to patients.
Conflict of interest. – The authors declare that there is no conflict of interest to indicate.
Funding. – This study was supported by the Natural Science Foundation of Liaoning Province (No. 2019-ZD-0598)
