Introduction
Salt stress and drought are major abiotic stressors that significantly affect all aspects of plant physiology, resulting in yield losses of more than 50% and a loss of more than $10.3 billion per year (Ma et al. 2020). Future global scenarios, envisioned by the Intergovernmental Panel on Climate Change indicate a decrease in precipitation and an increase in evapotranspiration rates (Pörtner et al. 2022). Initially, both stressors cause water deficit in plants, but under prolonged salinity, plants respond to hyper-ionic and hyper-osmotic stress in addition to dehydration (Chaves et al. 2009). Plant physiological responses to these stressors aim to minimize water deficit and restore ion homeostasis (Ma et al. 2020). Osmotic adjustments are achieved through the synthesis and accumulation of osmoprotective compounds (Chen and Murata 2002), while disturbances caused by excess sodium ions are remedied by changes in the activity and abundance of sodium/proton exchangers (Deinlein et al. 2014). In addition, both salinity and drought can induce the production of reactive oxygen species (ROS) that can cause lipid, protein, and DNA damage, which, however, can be counteracted by complex nonenzymatic antioxidants (glutathione, ascorbate, carotenoids) and antioxidant enzymes such as superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX) (Apel and Hirt 2004).
Transcriptomics studies in Arabidopsis revealed that 1008 and 1123 mRNAs are regulated in response to water deficit and salt stress, respectively (Apel and Hirt 2004), implying that both stresses involve complex processes. In sugar beet, an experiment with salinization and alkalinization identified 4773 and 2251 differentially expressed genes in leaves and roots, respectively (Geng et al. 2020). This and other studies identified ROS-scavenging enzymes, ion transporters and channels, proteins involved in signal transduction, and regulatory proteins, kinases, phosphatases, and transcription factors responsible for triggering the stress response (Zhu 2002; Yoshida et al. 2014). However, changes in mRNA levels do not correlate well with protein levels, and many gene products undergo posttranslational modifications that can alter protein activity (Varshavsky 1996; Deyholos 2010). Therefore, protein levels must be determined directly rather than extrapolated from transcript abundance. A complementary approach is to use proteomic analyses such as two-dimensional electrophoresis (2-DE) coupled with mass spectrometry (MS) to quantify protein abundance and identify it on a large scale (Hajheidari et al. 2005). To date, several papers have been published analysing the response to salt and drought stress using proteomic approaches (Hajheidari et al. 2005; Singh et al. 2022). Although there are common proteins that are regulated during salt and drought stress, each plant species has been shown to respond differently to these stressors. The differences between salt-tolerant (halophytes) and salt-sensitive (glycophytes) plants are particularly pronounced (Askari et al. 2006); (Zhang et al. 2012). Most proteomic research has focused on changes in the abundance of cellular proteins, while knowledge of stress-induced expression of extracellular proteins is limited. Only recently, it has been shown that the extracellular matrix and its constituent proteins are involved in the response to various stressors in rice, poplar and sweet potato (Zhang et al. 2009; Kim et al. 2013).
In this study we aimed to gain insight into the cellular and extracellular processes involved in responses to drought and salinity. To this end, we decided to use cells grown in vitro, since they represent a homogeneous system in which all cells are of similar origin and type, and the conditions of plant tissue culture allow the control of stress homogeneity and the characterization of cell behavior under stress conditions independently of the regulatory systems acting at the whole plant level (Errabii et al. 2007). On the other hand, plants are composed of a number of cell types that exhibit different cellular characteristics leading to different responses to stimuli. The N, HO, and HNO sugar beet cell lines have proven useful as in vitro models for studying epigenetic mechanisms, cell differentiation, and metabolism in plants (Le Dily et al. 1990; Causevic et al. 2006). The N line is a normal callus dependent on plant growth regulators, in contrast to the autonomous habituated HNO line and the tumorous T line, which is the result of cell transformation with the Agrobacterium tumefaciens Ti plasmid B6S3 (Pavoković et al. 2012b). In this study, we used the differentiated N line, which contains mainly parenchyma cells. It is photosynthetic and grows in response to 2,4-dichlorophenoxyacetic acid and 6-benzylaminopurine. It exhibits normal nuclear morphology and cell wall cellulose deposition (Pavoković et al. 2012b). To investigate the effects of salt- and mannitol-induced stress at the cellular level, we sought to identify stress-related proteins that are differentially expressed in non-stressed and stressed cells. We also determined possible changes in the concentrations of cellular macro- and microelements. In addition, this study was extended by analyzing the expression of stress-related extracellular proteins. We report the disruption of macro- and microelement homeostasis as a consequence of stress and the identification of novel proteins in sugar beet as stress-related proteins.
Plant material
Sugar beet N cell line ( Beta vulgaris L. subsp. vulgaris var. altissima Döll) was grown in vitro in modified Gamborg B 5 liquid nutrient medium (PG0; (Negrutiu et al. 1975; Pavoković et al. 2007). The growth chamber was maintained at 22 °C and a 16-h photoperiod (80 µmol photons m -2 s -1). Cells were subcultured every two weeks by transferring 10 mL of the old cells into 40 mL of fresh PG0 medium. The suspensions were shaken on a reciprocal shaker at 125 rpm.
Experimental conditions and harvesting
Salt stress was generated by growing cells in liquid PG0 medium containing 300 mM NaCl, while physiological drought was provoked by growing cells in the same medium containing 600 mM mannitol. Cells were harvested after 72 h of incubation, washed thoroughly with distilled H 2O, dried, and rapidly frozen in liquid nitrogen until use.
Macro- and microelements analysis
Five samples were used to determine macro- and microelement concentrations. Samples were analyzed by inductively coupled plasma atomic emission spectroscopy (ICP-AES) using the Prodigy High Dispersion ICP instrument (Teledyne Leeman Labs, Hudson, NH). ICP multi-element standard solution IV (Merck, Darmstadt, Germany) was used to control plasma positioning and to prepare standard solutions for calibration. All calibration standards were prepared by appropriate dilution of standard stock solutions (1 g L -1) in a concentration range from 0.1 to 5.0 mg L -1. Lyophilized samples were dried at a constant temperature of 70 °C for 1 h and then pulverized in a porcelain mortar. An amount of 0.15 g of each dried sample was weighed with analytical accuracy and placed in Teflon vials, except for the solutions of the nutrient medium. 4 mL of concentrated nitric acid, HNO 3, 1.0 mL of hydrogen peroxide, H 2O 2 (w = 30%) and 1.0 mL of ultrapure deionized water (R ≈ 18 MΏ) were added and the vials were left open for 30 min. The vials were sealed and placed in a rack holder of a microwave-assisted high-pressure digestion system (Berghof, Germany). Digestion was performed in several steps for 40 minutes. After cooling to room temperature, the solutions were filtered, transferred to 10 mL volumetric flasks, and filled to the mark with ultrapure deionized water. All samples were digested and analyzed as duplicates; blanks were also prepared in the same manner as the samples. To verify the accuracy of the digestion procedure, the same digestion scheme was applied to the certified reference material (SRM 1571 – Orchard leaves). The results are presented as mg macroelement per g of dry weight (DW) or µg microelement per g of DW together with the standard deviation of measurements.
Analysis of cellular proteins
The frozen cells were ground to a fine powder in liquid nitrogen using a pre-cooled mortar and pestle. For 2-DE, the phenol extraction protocol was performed according to a published procedure (Faurobert et al. 2007). Protein concentration was determined by the modified Bradford method using a UV/Vis spectrophotometer UV-4 (Unicam, UK) and bovine serum albumin (BSA) as a standard (Faurobert et al. 2007).
The first dimension, isoelectric focusing (IEF), was performed using 18 cm long nonlinear, immobilized pH gradient (IPG) strips, pH 3–10, in the IPGphor system (GE Healthcare, USA), according to Pavoković et al. (2012). The IPG strips were stored at -80 °C until use. IPG strips were thawed and incubated for 15 min in a buffer composed of 0.05 M Tris-HCl pH 8.8, 6 M urea, 2% SDS (w/v) containing 130 mM dithiothreitol (DTT) and then for 15 min in a buffer of the same composition, but with 135 mM iodoacetamide instead of DTT. The second dimension was performed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) as described in Pavoković et al. (2012a), using the PROTEAN II xi system (BioRad, USA).
Analysis of extracellular proteins
Extracellular proteins were harvested from the liquid medium after cells were removed. To remove debris, the medium was filtered through No 1. Whatman filter papers (Whatman, UK) and again through 0.45 µm Millipore filters (Millipore, USA). Proteins were concentrated on Amicon ultrafiltration devices (Millipore, USA) with the cut-off of 3 kDa. Protein concentration was determined by the Bradford method using a spectrophotometer and a BSA as standard (Bradford 1976). Proteins were mixed with Laemmli buffer (Laemmli 1970) and loaded onto a large vertical electrophoresis system. Electrophoresis was performed for 30 min at 100 V in stacking gel containing 4% T and 2.67% C and then at 220 V in running gel (12% T, 2.67% C) until the bromophenol blue ran off the gel.
Gel staining, image acquisition and analysis
Protein spots were visualized using Coomassie Brilliant Blue R-250 (CBB) or by silver staining (Blum et al. 1987). After protein visualization, gels were scanned with a flatbed scanner (HP, USA) at a resolution of 600 dpi and analyzed with Proteomeweaver 2.2 (Definiens, Germany) using the proposed working pipeline. Protein spots were detected using the following parameters: intensity limit, 10000; contrast limit, 50; and radius limit, 10. Statistical analyzes were performed using Proteomeweaver software. At least five gels, each from a biological replicate, were used to create the master gel for the control and for salt and mannitol treatments.
In-gel digestion, peptide sample preparation, and peptide mass spectrometry
Spots and bands of differentially expressed proteins were excised from the gels with a scalpel and the gel pieces were washed thoroughly using destaining buffer (30% methanol, 10% glacial acetic acid in H 2O) and prepared for matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry (MS) analysis as described in Pavoković et al. (2012b). Mass spectra were obtained using a MALDI-TOF/TOF MS (4800 Plus MALDI TOF/TOF analyzer, Applied Biosystems Inc., Foster City, CA, USA) equipped with a 200 Hz, 355 nm Nd:YAG laser, operated in the positive ion reflector mode.
For protein identification, we applied the global protein server explorer software (version 3.6, Applied Biosystems, USA) for Mascot (Matrix Science version 2.1, UK) search against the National Center for Biotechnology Information protein database (NCBIprot, http:// www.ncbi.nlm.nih.gov/protein).
Bioinformatics
The identified proteins were assigned to functional groups according to the MIPS Functional Catalogue FunCat database (Ruepp et al. 2004) and involved mechanisms reported previously in the literature. The target organelle of proteins was predicted using the TargetP web tool (Emanuelsson et al. 2007), which predicts whether protein sequences contain a mitochondrial target peptide, a chloroplast transit peptide or a signal peptide for secretion.
At the time of the initial experiment, mass spectrometry was used to identify protein sequences derived from organisms other than just sugar beet. To expand the disorder analysis to a set of proteins exclusive to this plant, we obtained sequences from the recently published sugar beet genome. We performed a similarity search of the initially identified protein sequences against the sugar beet genome assembly RefBeet-1.2 using the built-in BLAST within the Beta vulgaris Resource (http://bvseq.molgen.mpg.de/blast/) (Dohm et al. 2014).
The Universal Protein Resource (UniProt) was used for Gene Ontology analysis (GO, http://www.geneontology.org) of all identified proteins.
Statistical analysis
For the macro- and microelements analysis the results were analyzed with one-way ANOVA using the STATISTICA 13.0 (Stat Soft Inc., USA) software package. Differences between means were considered statistically significant at P < 0.05 (Tukey post hoc test for unequal sample sizes).
Statistical significance for protein expression in 2-D gels was calculated when the protein spot was present on at least three gels. Univariate mean differences between the abundance of protein spots in the control gel and in the gel obtained under salt or mannitol stress were examined using the non-parametric Mann-Whitney-Wilcoxon and Kolmogorov-Smirnov tests.
Disruption of homeostasis of macro- and microelements
Salt- and mannitol-induced stress resulted in disruption of cellular cation homeostasis (Tab. 1 and Tab. 2).
Tab.1. The concentration of macroelements in control cells of the sugar beet N cell line, in cells treated with 300 mM NaCl or 600 mM mannitol, and in modified Gamborg B 5 liquid nutrient medium (PG0) measured by inductively coupled plasma atomic emission spectroscopy. Results are mean values ± standard errors (n = 5). DW − dry weight.
Depending on the stress and element, different accumulation patterns were evident. The concentration of Na + ions was 7.7-fold higher in salt-treated cells than in the control but 8.3-fold lower with the mannitol treatment (Tab. 1). The concentrations of K + and Ca 2+ ions were decreased in both types of stress compared with the control, but this was particularly pronounced for K + ions in mannitol treatment (Tab. 1). In contrast, the stress did not affect the concentration of Mg 2+ ions. Both types of stress altered the concentrations of cellular microelements in different ways: NaCl decreased the content of Fe 2+, Cu 2+, and especially Mn 2+ ions and increased the concentration of Zn 2+ ions, whereas mannitol decreased the concentration of Fe 2+, Mn 2+, and Zn 2+ ions and increased that of Cu 2+ ions (Tab. 2).
Tab. 2. The concentration of selected microelements in control cells of the sugar beet N cell line, in cells treated with 300 mM NaCl or 600 mM mannitol, and in modified Gamborg B 5 liquid nutrient medium (PG0) measured by inductively coupled plasma atomic emission spectroscopy. Results are mean values ± standard errors (n = 5). DW − dry weight.
Salt- and mannitol-responsive cellular proteins
We applied a proteomic approach to analyze and compare the effect of salt and mannitol on the expression profile of sugar beet proteins. Salt stress resulted in statistically different regulation of 43 proteins, of which 22 were up-regulated and 21 were down-regulated (Fig. 1).
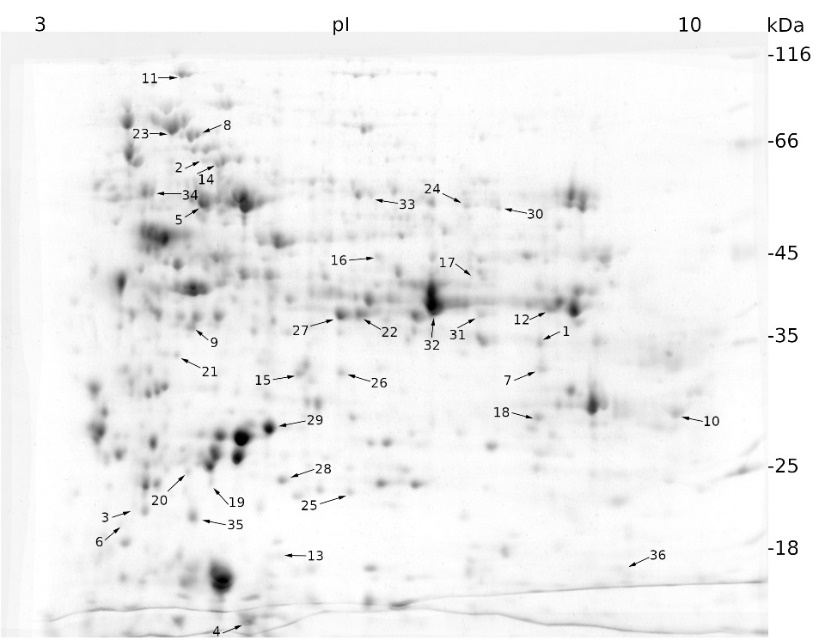
Fig. 1. Expression patterns of cellular proteins from a sugar beet N cell line exposed to salt stress (300 mM NaCl) for 72 h obtained by two-dimensional electrophoresis (2-DE). In the first dimension (IEF), 300 µg of proteins were resolved in 18 cm IPG strips with a non-linear pH gradient of 3-10. In the second dimension, proteins were separated on a 12% SDS polyacrylamide gel and then stained with Coomassie Brilliant Blue. Black arrows indicate proteins whose expression was statistically different to that of the control sample (see Materials and methods) and were identified using MALDI-TOF/TOF mass spectrometry.
Mannitol-induced stress affected the expression of 67 proteins, of which 41 were up-regulated and 26 were down-regulated (Fig. 2).
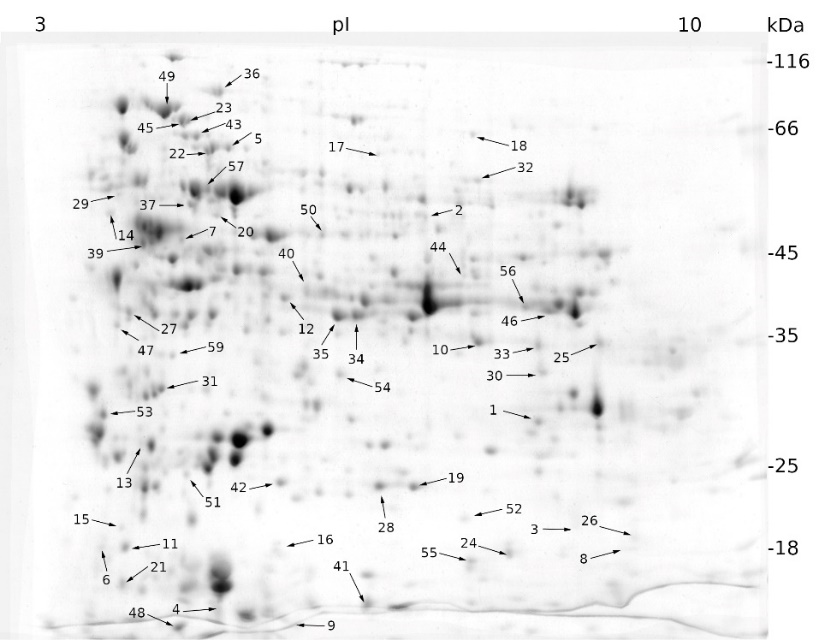
Fig. 2. Expression patterns of cellular proteins from a sugar beet N cell line exposed to mannitol stress (600 mM mannitol) for 72 h obtained by two-dimensional electrophoresis (2-DE) (for details see Fig. 1). Black arrows indicate proteins whose expression was statistically different to that of the control sample (see Materials and methods) and were identified using MALDI-TOF/TOF mass spectrometry.
Protein spots were excised and subjected to protein identification by MALDI-TOF/TOF MS. Of the proteins that were differentially expressed under salt stress, 25 were successfully identified, of which 15 were up-regulated and 10 were down-regulated (On-line Suppl. Tab. 1). Identification of proteins that showed differential expression after mannitol stress was successful for 29 of them, of which 18 were up-regulated and 11 were down-regulated (On-line Suppl. Tab. 2). Some salt- and mannitol-responsive spots were not excised due to their low abundance on the gel.
To obtain more information about unresolved proteins or those with low scores, we searched for protein homologs using the BLAST tool (Altschul et al. 1990). Functional classification of proteins was performed using the FunCat database (http://mips.helmholtz-muenchen.de/funcatDB/) (Ruepp et al. 2004). According to this classification, most of the identified proteins were involved in metabolism, energy and cell rescue, defense, and virulence, accounting for approximately 80% of the proteins (Fig. 3).
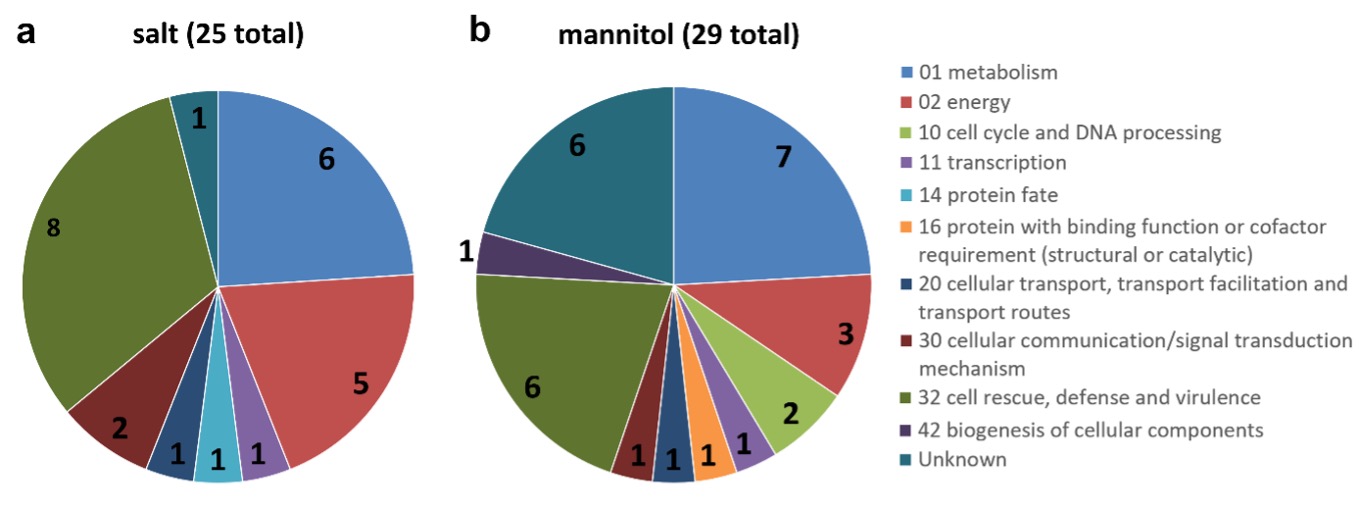
Fig. 3. Functional classification of the identified proteins: salt-induced stress proteins (a), mannitol-induced stress proteins (b). The functional grouping of the different proteins was based on FunCat web tool. The numbers on the pie-chart indicate the number of identified proteins belonging to the respective section.
GO analyses revealed that of the proteins that were up-regulated by salt stress, most were associated with the oxidative stress response (4 proteins) and the malate metabolic process (2 proteins), whereas other biological processes were represented by only one protein (Fig. 4a). Molecular function analysis revealed that most of the up-regulated proteins were related to hem-, ribosome-, and ATP-binding and to L-malate dehydrogenase activity (Fig. 4b) and exerted their function in the cytoplasm and chloroplast (7 and 4 proteins, respectively) (Fig. 4c). As for the down-regulated proteins, most of them are involved in the response to unfolded proteins (Fig. 4a) and have functions in ATP binding (Fig. 4b) in the cytoplasm (Fig. 4c).
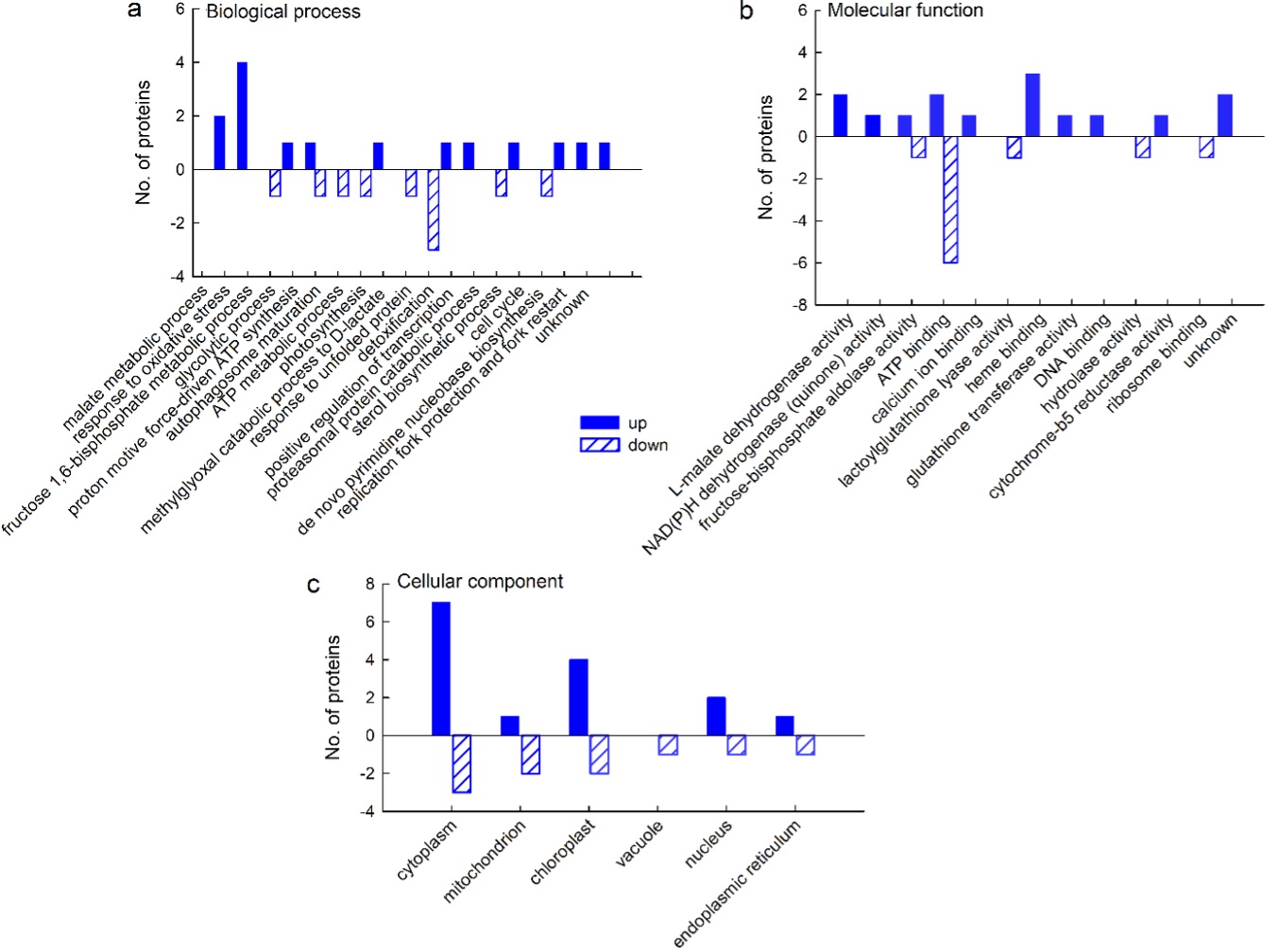
Fig. 4. Gene ontology (GO) analysis of cellular proteins from a sugar beet N cell line exposed to salt stress (300 mM NaCl) for 72 h: biological process (a), molecular function (b), and cellular compartment (c). GO analysis was derived through Uniprot hit accessions. Differently abundant proteins were identified by MALDI-TOF/TOF MS according to the NCBIprot database.
For most of the proteins that were up-regulated on mannitol, it was not possible to obtain information on the biological processes in which they are involved, but for those for which it was possible, GO analysis revealed that they are involved in malate metabolism and photosynthesis, as well as in the response to fungi and oxidative stress (2 proteins in each category) (Fig. 5a). Analysis of molecular function showed that two proteins had L-malate dehydrogenase activity, whereas other categories were represented by only one protein (Fig. 5b); the majority of the up-regulated proteins exerted their function in the cytoplasm (Fig. 5c). Of the down-regulated proteins, three are involved in glucose metabolism and two in protein folding and response to stress (Fig. 5a); their function is mainly in NAD and ATP binding (2 proteins per category) (Fig. 5b) in the cytoplasm and mitochondrial matrix (Fig. 5c), whereas other categories are represented by only one protein.
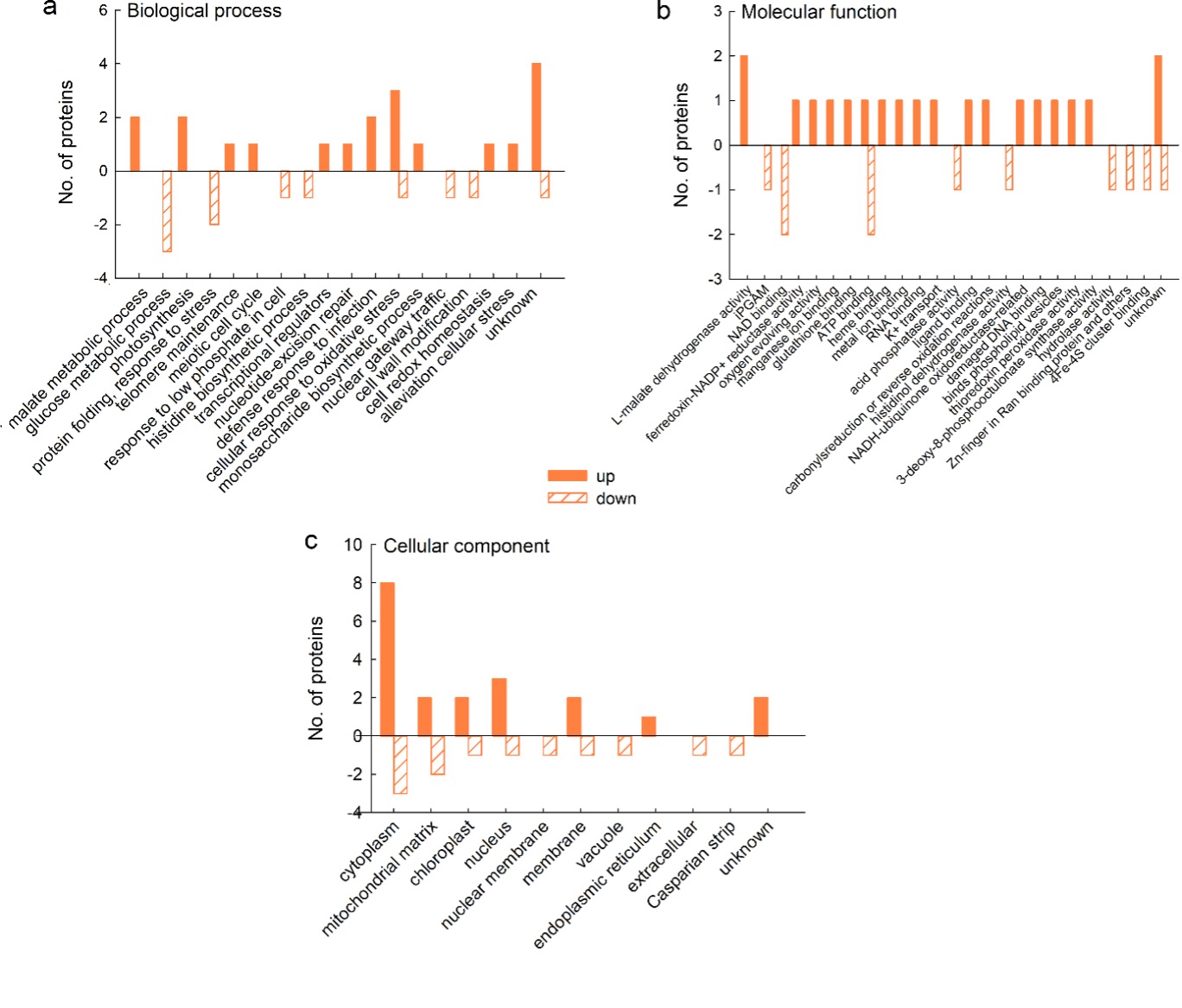
Fig. 5. Gene ontology (GO) analysis of cellular proteins from a sugar beet N cell line exposed to mannitol stress (600 mM mannitol) for 72 h: biological process (a), molecular function (b), and cellular compartment (c). GO analysis was derived through Uniprot hit accessions. Differently abundant proteins were identified by MALDI-TOF/TOF MS according to the NCBIprot database.
Salt- and mannitol-responsive extracellular proteins
Using experiments in cell culture, we investigated whether extracellular proteins are involved in the response to salt and mannitol stress. We found five protein spots (labelled E1-E5 in Fig. 6) whose expression changed significantly after salt and mannitol treatment compared with the control. They were excised from the gel and identified using MALDI-TOF/TOF MS.
Two of the five proteins with altered expression (labelled E1 and E4 in Fig. 6) were successfully identified. The other three proteins labelled E2, E3, and E5 are unknown (Fig. 6). Salt stress increased the expression of chitinase precursor (E4) and unknown proteins E2 and E3, whereas it down-regulated the expression of β-D-xylosidase 4 (E1) and unknown protein E5. The expression of β-D-xylosidase was also down-regulated by mannitol (On-line Suppl. Tab. 3).
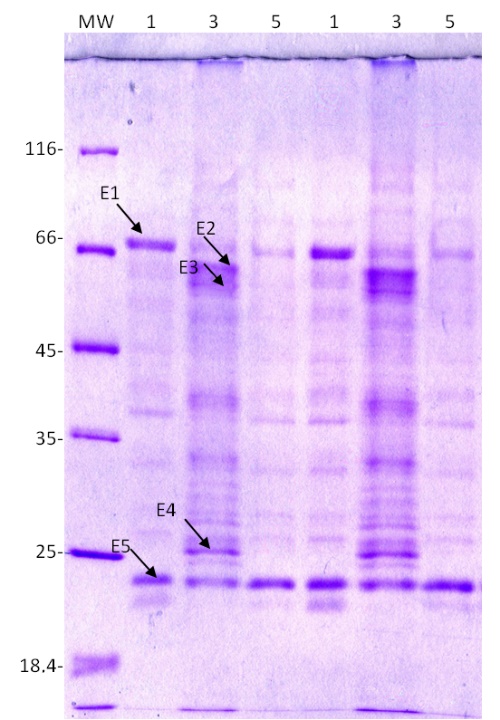
Fig. 6. Expression patterns of extracellular proteins from a sugar beet N cell line cultured for 72 h in modified Gamborg B 5 liquid nutrient medium (PG0) with or without the addition of 300 mM NaCl or 600 mM mannitol. Proteins were resolved on a 12% SDS polyacrylamide gel and stained with Coomassie Brilliant Blue. MW – molecular weight markers, 1 – control, 3 – 300 mM NaCl, 5 – 600 mM mannitol. E1-E5 represent proteins submitted to MALDI-TOF/TOF MS for identification.
GO analyses revealed that three proteins up-regulated by salt stress are involved in cell tip and root growth and carbohydrate metabolism and have a molecular function in binding copper ions and chitin. The proteins down-regulated by NaCl belong to arabinan catabolism, with alpha-L-arabinofuranosidase activity. The single mannitol-responsive protein that was down-regulated also belongs to arabinan metabolism, with alpha-L-arabinofuranosidase activity (Fig. 7).
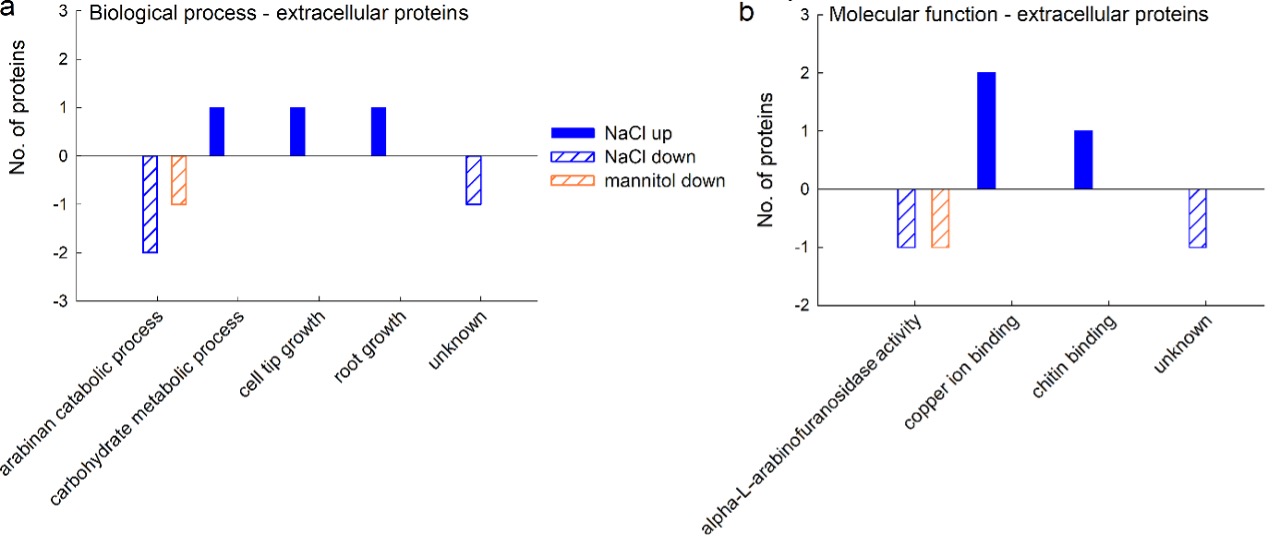
Fig. 7. Gene ontology (GO) analysis of extracellular proteins from a sugar beet N cell line exposed to salt (300 mM NaCl) and mannitol stress (600 mM mannitol) for 72 h: biological process (a) and molecular function (b). Differently abundant proteins were identified by MALDI-TOF/TOF MS according to the NCBIprot database. GO analysis was derived through Uniprot hit accessions.
Discussion
In this study, we investigated the cellular processes in sugar beet cells after a brief but severe exposure to salt and mannitol stress. Although sugar beet is an industrially important crop, there is little proteomic and bioinformatic information on the response of this plant to abiotic stress. By using cell lines instead of whole plants, our intention was to eliminate differences in protein expression between different tissues and focus on the effects of stress on a small number of cell types. We chose to use high concentrations of salt and mannitol to elicit a stronger cellular response. This approach allowed us to identify a number of previously unknown cellular and extracellular proteins involved in the response to salt and drought stress in sugar beet. A dose-dependent response to salt was also observed in the halophyte Suaeda aegyptica, where the number of regulated proteins was positively correlated with salt concentration (Askari et al. 2006).
Cation homeostasis
Salt and mannitol stress caused profound disruption of cation homeostasis in sugar beet cells. Salt-stressed cells increased the concentration of Na + ions, decreased the concentration of K + and Ca 2+, whereas the concentration of Mg 2+ ions did not change. Accumulation of Na + ions and loss of K + ions were observed in sugar beet treated with salt and sorbitol (Wu et al. 2014) and in Mammilaria gracilis callus and tumor exposed to NaCl (Balen et al. 2013). The loss of K + during salt stress appears to be related to antagonism of Na + and K + ions at uptake sites or inhibition of K + uptake at the plasma membrane (Hu and Schmidhalter 2005). This cellular uptake inhibition could lead to the measured lower Na + and K + levels and contribute to osmotic adjustment, in whole sugar beet seedlings during drought, for instance (Wu et al. 2014). Lower Ca 2+ levels can be explained by the displacement of extracellularly bound Ca 2+ ions by Na + ions and precipitation of Ca 2+ under saline conditions. Such Ca 2+ deficiency also allows passive uptake of Na + ions into cells, resulting in growth arrest and changes in morphology (Cramer et al. 1988). Loss of Ca 2+ has been observed upon salt stress in tomato plants and can be alleviated by small heat shock proteins (HSP) (Fu et al. 2016). Salinity affects micronutrients and nutrient solubility differently depending on the plant species (for a detailed discussion, see Hu and Schmidhalter 2005). It appears that high salinity reduces concentrations of these ions, as has been reported for shoots and roots of marigold (Koksal et al. 2016).
Mannitol decreased the concentration of all measured macroelements except Mg 2+. A strong decrease in the concentrations of Na +, K +, and Ca 2+ ions was also observed upon mannitol stress in M. gracilis callus (Balen et al. 2013). In addition, a small reduction in the concentration of K + and Na + ions and a large reduction in Ca 2+ ions were observed in various organs of sugar beet cv. Janus during long-term drought (Choluj et al. 2008). Ca 2+ concentration was reduced in shoots of two drought-sensitive cultivars during both mild and severe short-term drought (Wu et al. 2014). In addition, mannitol decreased Fe, Mn, and Zn microelement content but increased Cu content. The decrease in Fe, Mn, and Zn during drought is thought to be due to their lower solubility (Hu and Schmidhalter 2005). However, the decrease in accumulated ions under mannitol stress may be due to decreased ion uptake rather than the decreased ion mobility that occurs in soil-grown plants during drought (Choluj et al. 2008).
Proteome analysis
We compared the protein expression profile of control cells with that of cells under salt or mannitol stress. An approximately 1.3-fold difference in abundance compared with control was chosen as the threshold for protein significance, consistent with other similar proteomics studies (Parker et al. 2006; Yan et al. 2006; Jiang et al. 2007). We have identified proteins regulated by salt and drought stress such as H +ATPases (Kirsch et al. 1996), malate dehydrogenases (Song et al. 2001), heat shock proteins, oxidoreductases, and SOD, but also some new proteins that add to the available information on stress-related proteins in sugar beet. Classification using the FunCat tool showed that most of the identified proteins could be classified into three classes: (1) cell rescue, defense and virulence, (2) metabolism, and (3) energy.
Cell rescue, defense and virulence
ROS, such as singlet oxygen ( 1O 2), superoxide anion (O 2 •-) and hydrogen peroxide (H 2O 2), are formed in small amounts under normal growth conditions. Under abiotic stress, the production of ROS increases, which can cause damage to biological molecules (Hajheidari et al. 2005). To counteract ROS, plants employ families of enzymes that are part of their antioxidant system (Deyholos 2010). In the present study, a salt-dependent increase in protein abundance was observed for several APX isoforms and glutathione S-transferase (GST) 6, whereas mannitol increased protein abundance of the stromal APX isoform, GST F3 and Mn-SOD II. SODs catalyze the dismutation of O 2 •- to H 2O 2 H 2O 2 and O 2, and form the first line of defense against the toxic effects induced by ROS (Salekdeh et al. 2002). H 2O 2 is then detoxified to H 2O by the APX family, which consists of several isoforms localized in thylakoid and glyoxisome membranes as well as in the chloroplast stroma and cytosol (Gill and Tuteja 2010). In addition, H 2O 2 H 2O 2 is degraded by peroxiredoxins, and the isoform up-regulated in this study is targeted to chloroplasts, suggesting that ROS production was increased in this organelle (Dietz 2007). Endogenous and xenobiotic toxic compounds can be detoxified by GSTs, which form a large family consisting of forty-seven members identified in the Arabidopsis genome (Gill and Tuteja 2010). The GSTs up-regulated in this study appear to belong to the plant-specific Type I (phi-class), whose function is to counteract oxidative damage induced by herbicides and abiotic stress (Wagner et al. 2002; Gill and Tuteja 2010).
Salt- and mannitol-induced stress regulates another large group of proteins: the family of heat shock proteins (HSPs). In plants, HSPs are classified into five families according to their molecular weight, amino acid sequence homology, and functions: HSP100, HSP90, HSP70, HSP60, and the small HSP. They can be up-regulated under abiotic stress to support proper folding, unfolding and translocation of proteins to target organelles (Gupta et al. 2010). In this study, the chloroplast and mitochondrial HSP isoforms, together with a chloroplast 60 kDa chaperonin subunit, were significantly down-regulated after salt- and mannitol-induced stress, which may be due to the lower protein synthesis rate as reported in barley roots after salt shock (Hurkman and Tanaka 1987). Down-regulation of chloroplast HSP70 and HSP90 isoforms has also been reported during salt stress in Camellia sinensis (Wang et al. 2015; Chen et al. 2018). Moreover, HSP70 was down-regulated in callus and tumor tissues of M. gracillis treated with either NaCl or mannitol, while HSP60, which is critical for achieving native forms of newly synthesized proteins, was also down-regulated in tumor exposed to NaCl (Rogić et al. 2015). HSP70 can inhibit the transcription of other HSPs by binding to the heat shock transcription factor (HSF) (Kim and Schöffl 2002; Oliver et al. 2011). Reducing HSP70 levels allows for increased production of small HSPs, which play an important role in protection against drought and salt stress (Zou et al. 2012).
Glyoxalase I (lactoylglutathione lyase) was down-regulated in sugar beet cells under salt stress. It is an enzyme involved in the detoxification of the cytotoxic compound methylglyoxal (MG), which is a byproduct of glycolysis. This enzyme was also down-regulated in tobacco cells under salt stress (Hoque et al. 2008), suggesting that detoxification of MG via the glyoxalase system was insufficient in cells under salt stress. Another possible explanation could be that the metabolism of triose phosphates and thus the production of MG was reduced in cells under salt stress.
Metabolism
At the physiological level, salt stress and drought decrease stomatal conductance and photosynthesis, and increase photorespiration (Atkin and Macherel 2009). During detoxification of photorespiration products, there is an increased requirement for the oxidation of NADH and regeneration of NAD +, which occurs in mitochondria using the mitochondrial NADH dehydrogenase type II or the malate/oxaloacetate shuttle across the inner mitochondrial membrane (Atkin and Macherel 2009). A deficiency of NAD + in the cell is detrimental because an adequate amount of ATP cannot be provided during stress. Therefore, the increased abundance of cytoplasmic and mitochondrial malate dehydrogenases detected in our study during salt- and mannitol-induced stress serves to ensure ATP and support normal chloroplast function (Liu et al. 2012). An increase in malate dehydrogenase levels was also observed under 150 mM NaCl stress in Arabidopsis, under treatment with 300 mM NaCl in Eremochloa ophiuroides, and during drought in Elymus elongatum (Jiang et al. 2007; Salekdeh et al. 2007; Liu et al. 2012).
We observed up-regulation of one isoform of fructose-biphosphate aldolase and down-regulation of another isoform, aldolase superfamily protein, after salt stress. In plants, there is a cytosolic and a chloroplast isoform of this enzyme (Konishi et al. 2004). We performed protein localization estimation based on the target peptide sequence using the TargetP algorithm (Emanuelsson et al. 2007), and the cytosolic isoform was up-regulated, whereas the chloroplast isoform was down-regulated. Up-regulation of the cytoplasmic isoform of the enzyme appears to help plants to cope with anaerobic conditions by promoting the function of the glycolytic pathway for ATP synthesis (Konishi et al. 2004). The down-regulation of the chloroplast isoform is likely a consequence of lower photosynthetic rates and the establishment of tolerance following stress induction (Yamada et al. 2000).
Salt stress resulted in up-regulation of putative quinone reductase, an enzyme belonging to the large family of oxidoreductases that acts on NADH or NADPH with a quinone or similar compound as acceptor. It appears to be involved in the detoxification of reactive carbonyls in plants (Yamauchi et al. 2010). Quinone reductase was also found to be up-regulated after salt stress in tomato (Zhou et al. 2009). We also observed mannitol-induced down-regulation of histidinol dehydrogenase, which is part of amino acid metabolism and catalyzes the final step in the production of histidine from histidinol. Down-regulation of this enzyme has also been reported in Chlamydomonas reinhardtii exposed to heat shock, suggesting that a reduction in protein biosynthesis occurs during abiotic stress, perhaps as a means to reduce problems with de novo protein folding under adverse conditions (Mühlhaus et al. 2011).
Energy
Four different proteins belonging to ATPases were differentially regulated after salt stress: proteins belonging to the endoplasmic reticulum (ER), mitochondria, and V-type ATPases were down-regulated, whereas a subunit of the chloroplast ATPase was up-regulated. Transitional ER ATPase provides energy for vesicle budding, which is responsible for transport between ER and Golgi (Zhang et al. 1994). Down-regulation of this enzyme after salt stress could be a consequence of decreased protein synthesis, which then leads to decreased vesicle production and protein transport. V-type and mitochondrial ATPases are important in salt stress to stimulate the transport of Na + ions in the vacuole and to restore ion balance, respectively (Lehr et al. 1999; Ndimba et al. 2005). However, the beta-subunit of mitochondrial ATP synthase was down-regulated in rice roots under severe salt stress (500 mM for 2 h), which ultimately triggered programmed cell death in salt-sensitive rice (Chen et al. 2009).
Salt stress up-regulated two proteins involved in photosynthesis: a β-subunit of chloroplast ATP-synthase (AtpB) and a 23 kDa polypeptide of the oxygen-evolving complex (OEC) (PsbP) of the photosystem II (PSII), whereas mannitol up-regulated the 33 kDa OEC protein (PsbO). PsbP and PsbO are extrinsic proteins located on the luminal side of the PSII complex in chloroplasts, where they serve to maintain oxygen evolution at physiological rates and ensure thylakoid stability (De Las Rivas et al. 2007). Their increase under salt stress could help maintain sufficient water-splitting activity or stabilize PSII under these conditions (Yamauchi and Sugimoto 2010). Up-regulation of PsbP protein has also been observed in Norway spruce needles under mild drought conditions (Blödner et al. 2007).
Mannitol up-regulated ferredoxin-NADP + reductase (FNR), an enzyme that catalyzes the final step of electron transfer in PSI and is responsible for NADPH production. A general increase in FNR transcripts was observed in Arabidopsis plants exposed to drought stress (Lehtimäki et al. 2010), and it was concluded that FNR is important in redox regulation and in antioxidant mechanisms in chloroplasts during drought stress.
Mannitol also up-regulated mitochondrial NADH-ubiquinone oxidoreductase, also known as Complex I of the respiratory chain. The complex transfers electrons from NADH to coenzyme Q and translocates protons across the inner mitochondrial membrane, contributing to the buildup of the electrochemical potential needed to produce ATP (Brandt 2006). The up-regulation of the enzyme during drought could help mitochondria to restore ROS imbalance after stress (Nwugo and Huerta 2011; Sharma et al. 2011).
Mannitol stress resulted in down-regulation of the glycolysis enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (iPGAM). GAPDH, which catalyzes the reversible oxidative phosphorylation of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate, requires NAD(P)H for its activity and is normally inhibited when cells are exposed to oxidative stress (Chernyad’ev and Monakhova 2006; Ralser et al. 2007). IPGAM was found to be down-regulated in Thellungiella halophila upon salt stress, likely due to decreased energy metabolism and subsequent formation of ROS (Gao et al. 2008). Moreover, expression of both enzymes was also reduced in M. gracilis tumor exposed to salt and mannitol (Rogić et al. 2015). Taken together, these results suggest that primary metabolism in exposed tissues was significantly affected by salt- and mannitol-induced stress and that, in response to salinity and osmotic stress, energy metabolism is decreased to reduce the excessive production of ROS that could trigger oxidative stress (Gao et al. 2008).
Other categories of proteins
The proteasome is a large protein complex that eliminates damaged or misfolded proteins in cells. Here, we found down-regulation of the α-subunit of the proteasome 20S complex in response to salt stress. Considering that stress increases the number of misfolded and damaged proteins, one would expect an increase in 20S abundance. However, in wheat roots, 20S proteasome subunit abundance decreased under salt stress, although activity increased (Shi et al. 2011), suggesting that regulation of proteasome activity is not always well correlated with the stress severity. In addition, the 26S proteasome subunit α was found to be down-regulated in M. gracillis tumor tissue treated with NaCl (Rogić et al. 2015), and the authors speculated that under stress, plants process misfolded proteins mainly by refolding. Proteasome abundance may also be down-regulated during programmed cell death (Kurepa and Smalle 2008).
The down-regulation of receptors for activated C kinase 1 (RACK1) by salt stress appears to be related to its regulation by the stress hormone abscisic acid (ABA). RACK1, considered a versatile scaffold protein, is a negative regulator of ABA responses. Induction of ABA during salt and drought stress down-regulated the expression of three RackK1 isoforms in Arabidopsis (Guo et al. 2009a). ABA can modulate gene expression depending on whether or not new protein synthesis is required (Zhang et al. 2006). Group A of the bZIP family of transcription factors, the ABRE-BINDING PROTEIN/FACTOR (AREB/ABF) family, is responsible for gene expression that has a ABA-responsive cis-element (ABRE) in its promoter domain (Kim et al. 2011). The bZIP transcription factor up-regulated in this study may be a member of the AREB family in sugar beet responsible for mediating ABA responses.
Mannitol up-regulated a glycine-rich RNA-binding protein. The superfamily of glycine-rich proteins is involved in post-transcriptional processes such as mRNA and rRNA processing, RNA export, and stability (Hu et al. 2011). A β-subunit of a probable voltage-gated potassium channel localized in the plasma membrane was strongly up-regulated by mannitol. In mammals, β-subunits are not directly involved in potassium transport but in modulating channel activity and are stoichiometrically related to α-subunits transporting ions according to the formula α4β4 (Capera et al. 2019). Ardie et. al. (Ardie et al. 2011) used a β-subunit of a K + channel from the halophyte Puccinellia tenuiflora and induced it transiently in yeast and Arabidopsis. The activity of the protein can alter the levels of K + and Na + in plant parts. During drought, cells suffer from a deficiency of K + ions (Hu and Schmidhalter 2005) and plants try to maintain a high K +:Na + ratio to ensure the integrity of cell membranes. These results suggest that in sugar beet cells, as in mammals, β-subunits may be involved in modulating K + channel activity during stress.
Extracellular proteins
In this study, we observed only five extracellular proteins with altered expression. Although the number is lower than reported in other studies (Zhang et al. 2009, Pechanova et al. 2010), this is the first analysis of this type in sugar beet. A β-xylosidase was down-regulated during salt- and mannitol-induced stress. This enzyme belongs to a family of exo-hydrolases but also has significant transglycosylase activity (Franková and Fry 2011). It is involved in the degradation/reconstruction of xylose-containing polysaccharides in the plant cell wall during various physiological events (Franková and Fry 2011). The down-regulation of the enzyme during elevated salinity and drought appears to be part of a defense mechanism in which no cell wall modification serves to alleviate cell damage.
AT1G76160 [ Arabidopsis thaliana] L-ascorbate oxidase (SKU5 similar protein 5, SKS5), hypothetical protein At1g41830 (SKU5 similar proteins 6, SKS6), and a chitinase were up-regulated only during salt stress. SKS5 and SKS6 belong to a family of multicopper oxidase-like proteins related to ferroxidases, ascorbate oxidases, and laccases (Jacobs and Roe 2005). SKS6 is involved in the formation of vascular patterns in cotyledons during Arabidopsis development, and is positively influenced by plant hormones such as ABA, 1-aminocyclopropane-1-carboxylic acid (ACC, the direct precursor of ethylene), indole-3-acetic acid (IAA, the most common naturally occurring plant hormone of the auxin class), and 2,4-dichlorophenoxyacetic acid (2,4-D, a synthetic auxin) (Jacobs and Roe 2005). The role of SKS5 in the cell wall is not known but may be related to hyperhydric stress (Sen and Alikamanoglu 2013). The chitinase protein has a domain of glycoside hydrolase 19 (GH19) and belongs to Class IV chitinase by size. While the primary role of this class of chitinases is in defense against pathogens, up-regulation at the transcriptional level has been observed in Arabidopsis under salt and drought stress (Takenaka et al. 2009; Vaghela et al. 2022).
In conclusion, we showed that both stressors altered ion status in cells. Most of the regulated proteins were responsible for alleviating the increased ROS production and cellular damage and restoring homeostasis. In addition to previously known cellular proteins commonly identified as stress-responsive, we have identified several new proteins that appear to be involved in the stress response. Their role in abiotic stress in sugar beet requires further investigation. By extending our study to the extracellular space, we also identified several extracellular proteins involved in the stress response.
Acknowledgments
This work was supported by a grant no. 119-1191196-1200 to prof. dr. sc. Marijana Krsnik-Rasol, a grant “Life under stress: molecular components and mechanisms of plant response to drought and salinity stress” to prof. dr. sc. Marijana Krsnik-Rasol and prof. Dudy Bar-Zvi and a grant “Mechanisms of plant defenses to abiotic stress: changes of proteome due to salt and osmotic stress” to prof. dr. sc. Biljana Balen. Grants are from the Ministry of Science and Education of the Republic of Croatia and University of Zagreb, Croatia.
