Introduction
The biotechnological application of lactic acid bacteria (LAB) in food, and in the dairy industry is continuous. In this context, also an important physiological property of certain LAB is the proteolytic activity. The proteolytic system of LAB contributes to the organoleptic properties of fermented dairy products (Blaya et al., 2018; Beganović et al., 2013; Leboš Pavunc et al., 2012), but also has potential due to the synthesis of bioactive peptides, also known as biopeptides, in probiotic strains. For example, Waśko et al. (2012) described purification of serine proteinase of Lacticaseibacillus rhamnosus OXY isolated from human gastrointestinal tract. Namely, the proteolitic breakdown of casein results in the accumulation of the amino acids, necessary for growth in milk (Villegas et al., 2015; Waśko et al., 2012). LAB require a variety of amino acids for growth, whose availability in the cheese microenvironment is low (Brown et al., 2017). Proteolytic enzymes supply LAB cells with nitrogen compounds. Therefore, the primary function of enzymes of the proteolytic system is the breakdown of proteins into peptides, or amino acids that can be utilised as sources of nutrients (Kieliszek et al., 2021). In addition, the proteinase activity of certain LAB strains can result in the synthesis of bioactive peptides in dairy products. These peptides can be formed by hydrolysis of food proteins through the action of digestive enzymes, or by in vitro enzymatic hydrolysis and microbial fermentation (Marcone et al., 2017). The potential of bioactive peptides is also significant within the concept of healthy eating due to their beneficial effects (Huang and Kok, 2020). When trapped in the casein structure, they are intact and not biologically active. When liberated, their beneficial effects are multifunctional and include role in mineral absorption, antihemolytic, antimutagenic, antioxidant, immunomodulatory or antimicrobial activities (Raveschot et al., 2018; Aguilar-Toalá et al., 2017; Marcone et al., 2017). Certain L. helveticus strains have the potential to produce peptides Val-Pro-Pro and Ile-Pro-Pro in fermented dairy products, which are associated with antihypertensive effects (Cremonessi et al., 2013). Bioactive peptides have been found in different cheeses (Raveschot et al., 2020; Lim et al., 2017; Golić et al., 2013). Namely, according to Golić et al. (2013) 44.3 % out of 87 LAB strains from white-pickled and fresh soft cheeses showed proteolytic activity when incubated with casein. Some of these LAB, which were isolated from Croatian fresh soft cheeses and identified as Lc. lactis, were also included in this study. The aim of this study was to examine the proteolytic activity of several Lactobacillus and Lactococcus autochthonous strains with a fast milk-coagulating (Fmc +) phenotype which are isolated from artisanal fermented foods, mainly cheeses, from Croatia. In parallel the pH values and percentage of lactic acid were monitored to assess potential of tested LAB strains to acidify the milk.
The possibility of inducing proteinase activity of producer strains was also examined by inoculating strains in skimmed milk instead of optimal medium (MRS for Lactobacillus sp. or M17 for Lactococcus sp.). Following the in vitro selection of potential proteinase producer strains, extraction of putative proteinases was performed, which were then analysed by the SDS-PAGE method.
Materials and methods
Bacteria and culturing conditions
Lactobacillus and Lactococcus strains, from the Culture Collection of the Laboratory of Antibiotic, Enzyme, Probiotic and Starter Culture Technologies, Faculty of Food Technology and Biotechnology, University of Zagreb (CIM-FFTB) Zagreb, used in this study, are listed in Table 1. These LAB strains are isolated from different ecosystems of fermented products, mostly from autochthonous Croatian cheeses.
Table 1. Lactobacillus and Lactococcus lactis strains tested for proteolytic activities
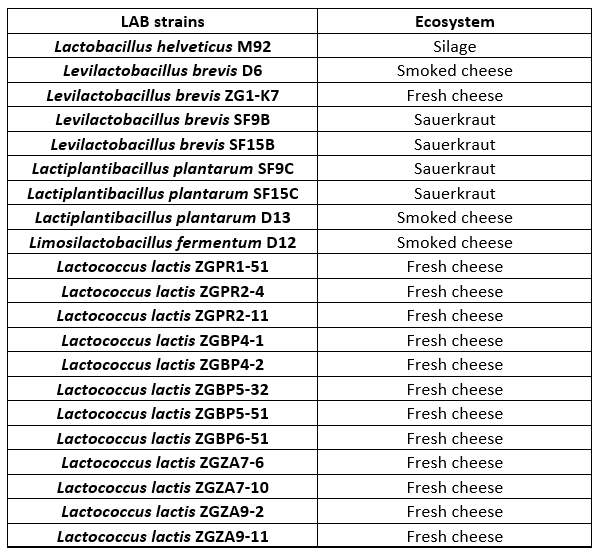
LAB strains are maintained as frozen stocks at -80 °C in MRS (BioLab ZRt, Budapest, Hungary) or M17 broth (Biolife, Milano, Italy), respectively, with the addition of 15 % (by volume) glycerol (Alkaloid, Skopje, North Macedonia). Prior to the experiments, Lactobacillus strains were routinely propagated in MRS broth, while Lactococcus strains in M17 broth and incubated overnight at 37 °C in anaerobic conditions.
Milk coagulation test
The coagulation capacity of LAB strains was determined as essentially described by Hebert et al. (2001). LAB cells, propagated in MRS at 37 °C overnight, were harvested by centrifugation at 8000 x g for 10 min at 4 °C, and washed twice in sterile 50 mM sodium phosphate buffer (pH 7.0). This cell suspension was used to inoculate 10 % (m/V) reconstituted skimmed milk (RSM) and were incubated at 37 °C for 16 h. LAB strains that showed Fmc + phenotypewere indicated as strains with excellent coagulation capacity (+++), while other obtained results were indicated as: no coagulation (–), poor coagulation (+/-), moderate coagulation (+) or good coagulation capacity (++).
Qualitative determination of proteolytic activity
The proteolytic activity of the LAB was qualitatively detected after overnight growth in optimal medium or RSM, using the skimmed milk agar as described by Raveschot et al. (2020) with slight modifications. A 50 l of LAB biomass after centrifugation of 50 µl of cell supernatant was placed on agar containing 1 % ( m/V) skimmed milk (MerckKGaA, Darmstadt, Germany) and incubated at 30 °C for 48 h. Proteolytic activity was indicated by the occurrence of a clear hydrolysis zone. All analyses were performed in triplicates.
Selected strains, that showed caseinolytic activity, were inoculated with the vertical line test on skimmed milk with 1 % ( m/V) agar. After 48 h of incubation, transparent zones around the bacterial growth were photographed.
pH values and titratable acidity ( oSH)
pH values and titratable acidity ( oSH) were determined after overnight growth of Lactobacillus and Lactococcus strains in optimal medium (MRS or M17 broth) and skimmed milk with three repetitions, as previously described by Leboš Pavunc et al. (2009).
Extraction of proteinase
Extraction of proteinase was performed as previously described by Villegas et al. (2015) with slight modifications. Lactococcus and Lactobacillus cells from the exponential growth phase were harvested by centrifugation at 8000×g for 10 min at 4 °C, washed twice with 0.85 % ( m/V) saline supplemented with 10 mM CaCl 2 (Gram-Mol, Zagreb, Croatia), and resuspended to in 20 mM Tris–HCl buffer of pH 7.5 (Invitrogen, Carlsbad, USA) containing 5 mM CaCl 2. Samples were incubated for 30 min at room temperature and then centrifuged at 8000×g for 15 min. The supernatants of protein extracts were stored at 4 °C, and the precipitated protein pellets were washed, resuspended in 20 mM Tris–HCl buffer and stored at 4 °C. The protein concentration was measured by a BioSpec-nano-spectrophotometer (Shimadzu, Japan). Supernatants of protein extracts and protein pellet fractions were subjected to proteolytic activity and SDS-PAGE analyses. In order to further concentrate and purify proteinase, ultrafiltration of supernatants of 7 selected strains, obtained after proteinase extractions, was performed by filtration through a 100-kDa membrane (100,000 MWCO, Amicon, Millipore) at 4000 g during 10 min, according to the manufacturer’s instruction, yielding a partially purified proteinase sample.
SDS-PAGE analysis
SDS-PAGE was performed according to the method of Laemmli (1970). Resolving and stacking gels contained 8 and 4 % ( m/V) acrylamide (MerckKGaA, Darmstadt, Germany), respectively, and the samples were treated by boiling for 2 minutes with reducing reagents. Electrophoresis was performed at 50 V for 3 h. ProSieve ®QuadColor™ Protein Marker for SDS
Electrophoresis containing 12 proteins ranging in size from 4.6 to 315 kDa, was used as a molecular weight marker. The proteins were visualized by staining with 0.02 % Coomassie Brilliant Blue G-250 (MerckKGaA, Darmstadt, Germany), 25 % (by volume) isopropanol (Kemika, Zagreb, Croatia) and 10 % (by volume) acetic acid (Carlo Erba Reagents, Cornaredo, Italy) during 3 hours followed by destaining in 10 % (by volume) of acetic acid. Gel images were digitised with HP Photosmart 5510 Scanner.
Statistical analyses
Statistical analyses were performed with the Statistical Computation Web Site „VassarStats“ using the Tukey’s post hoc test, and P<0.05 was considered significant. Experiments were performed in triplicate. All reported values were the means of three independent trials ± standard deviation.
Results and discussion
Screening for caseinolytic LAB
Caseinolytic potential of a selection of LAB strains previously isolated in Croatia is described, including Lc. lactis strains from autochthonous fresh cheeses and probiotic or functional starter Lactobacillus strains from fermented foods. Lc. lactis and certain Lactobacillus strains are considered as the most important industrial dairy starter cultures (Chlebowska-Smigiel et al., 2017; Leboš Pavunc et al., 2012). LAB proteinases besides role in cell growth and aroma formation, are even capable of hydrolysing allergenic proteins in milk (Kieliszek et al., 2021).
Proteinase expression in some Lactococcus and Lactobacillus strains may be associated with the incidence of the fast milk coagulating (Fmc +) phenotype, or even with acidifying capacity as reported for Streptococcus thermophilus (Dandoy et al., 2011; Herbert et al., 2001). Moreover, certain LAB are biopeptide producers, albeit in lower concentrations which may contribute to the functional value of fermented dairy products (Yusuf and Gürkan, 2021; Marcone et al., 2017; McSweeney and Fox, 2017; Venegas-Ortega et al., 2019; Raveschot el., 2018). Biopeptide synthesis by LAB is attractive from another aspect to a dairy industry because the bioprocess contributes to the quality of the final product without need for additional technological procedures and costs (Ravenschot et al., 2020; Brown et al., 2017). Efforts are already being made to increase the biopeptides synthesis by genetic manipulations of the proteolytic system of the Lactococcus lactis MG1363 (Huang and Kok, 2020). Hansen and Marcatili (2020) predicted the structure of cell envelope proteinase and the interactions of Lc. lactis cells with casein. Therefore, due to the recognized importance of LAB proteases, we were interested in estimating the frequency of proteolytic strains among LAB population of fermented food products.
For a preliminary assessment of the use of LAB strains as potential starter cultures for cheese fermentation, the possibility of 1 % inoculum to coagulate milk during overnight incubation at 42 °C was observed, by determining the Fmc + phenotype (Table 2). A selection of 21 Lactobacillus or Lc. lactis strains, isolated from different food fermentations, primarily fermented dairy products, were used in the analysis. Certain LAB strains were previously characterised, such as L. fermentum D12, an exopolysaccharide producer (Butorac et al., 2021) or S-layer-carrying L. brevis SF9C and plantaricin-producing L. plantarum SF9B (Butorac et al., 2020; Banić et al., 2018) and S-layer expressing L. brevis D6 (Uroić et al., 2016). Overall, Lc. lactis strains efficiently coagulated skimmed milk. Among Lactobacillus strains, L. helveticus M92, L. brevis D6 and ZG1-K7 were effective in skimmed milk coagulation as observed by appearance of Fmc + phenotype (Table 2).
Table 2. Qualitative detection of coagulation and the presence of Fmc + phenotype after 16 h cultivation of Lactobacillus and Lactococcus strains in skimmed milk. The results represent three individual repetitions, indicated as: – no coagulation; +/- poor coagulation; + moderate coagulation; ++ good coagulation; +++ excellent coagulation
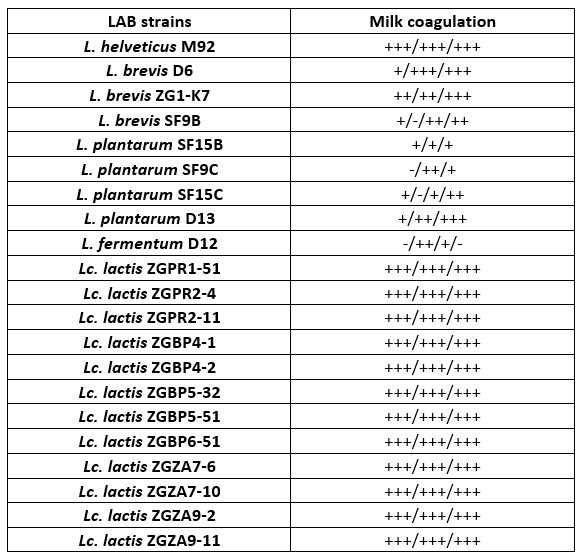
Coagulation is a crucial process in cheesemaking that occurs simultaneously with acidification. The synthesis of high concentrations of lactic acid by dairy starter cultures determines besides organoleptic characteristics, also cheese quality (Villegas et al., 2015; Leboš Pavunc at al., 2013). More precisely, the growth of the cheese spoilage bacteria is inhibited by acidification. The rate of milk acidification by Lactococcus and Lactobacillus starter cultures is therefore of major technological importance. During fermentation, the efficient growth of LAB strains is manifested by the conversion of lactose into lactic acid, which results in a lowering the pH of the medium. Therefore, the pH values and the titratable acidity in the culture supernatants were determined (Table 3 and 4). When comparing the titratable acidity among the strains, L. plantarum D13 after the overnight growth in the optimal medium and L. helveticus M92 after the overnight growth in skimmed milk showed the fastest acidifying potential. L. plantarum D13 is a probiotic starter culture isolated from smoked fresh cheese (Uroić et al., 2016), while L. helveticus M92 is a proteolytic strain (Beganović et al., 2013). The rapid growth of L. helveticus strains in milk is explained by pronounced activity of the proteolytic system and adaptation to low pH values (Blaya et al., 2018; Griffiths and Telez, 2013; Beganović et al., 2013; Cremonesi et al., 2013). Generally, Lactobacillus strains significantly lowered the pH values compared to Lactococcus strains, when they were grown in optimal medium (Table 3). Conversely, after cultivation in skimmed milk, Lc. lactis strains significantly lowered the pH values of the supernatant compared to L. fermentum D12 and L. plantarum strains (Table 3).
Table 3. pH values determined in the culture supernatants after overnight cultivation of Lactobacillus and Lactococcus strains in optimal medium or skimmed milk
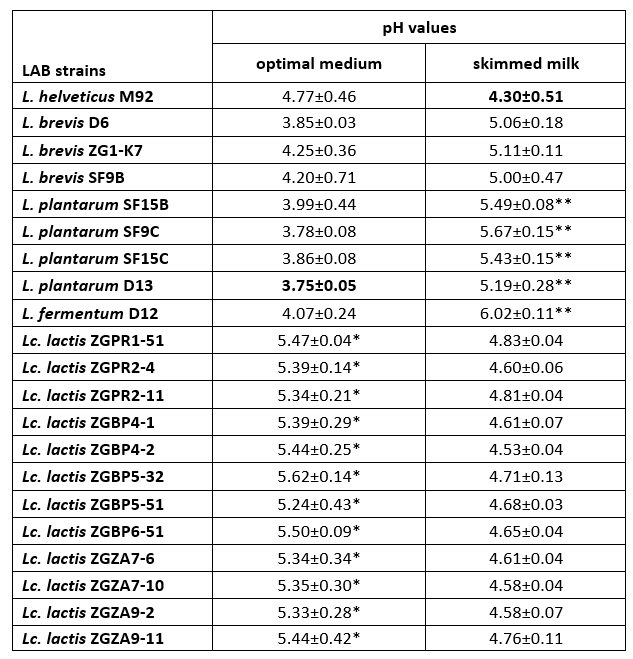
*The pH values of the culture supernatants, statistically significantly different (p<0.1) from the lowest pH value determined in the culture supernatant of L. plantarum D13 after overnight cultivation in optimal medium.
**The pH values of the culture supernatants, statistically significantly different (p<0.5 for SF15B and D13 strain; p<0.1 for SF9C, SF15C and D12 strain) from the lowest pH value determined in the culture supernatant of L. helveticus M92 after overnight cultivation in skimmed milk
The increase in titratable acidity was accompanied by a decrease in the pH of culture supernatant of each LAB strain. The highest titratable acidity was measured for L. helveticus M92 after overnight cultivation in skimmed milk (Table 4). Previous researches have highlighted a link between the presence of a casein proteolytic system and the fast growth and acidification rate of LAB in milk (Dandoy et al., 2011; Heberet et al., 2001). We have showed that all of Lc. lactis strains have ability to coagulate milk. In Lactococcus, the conversion of an Fmc + to an Fmc - phenotype has mainly been attributed to loss of either a PrtP proteinase or the enzymes for lactose utilization. Hebert et al. (2001) proved that the loss of a plasmid encoding an oligopeptide permease system influenced the transformation of the fast Lc. lactis to the slow milk coagulating strain. If any of these traits are not present, lactic acid production capacity is also lower and the Lactococcus and Lactobacillus strains are no longer effective starter cultures.
Table 4. Titratable acidity (°SH) determined in the culture supernatants after overnight growth of Lactobacillus and Lactococcus strains in optimal medium or skimmed milk
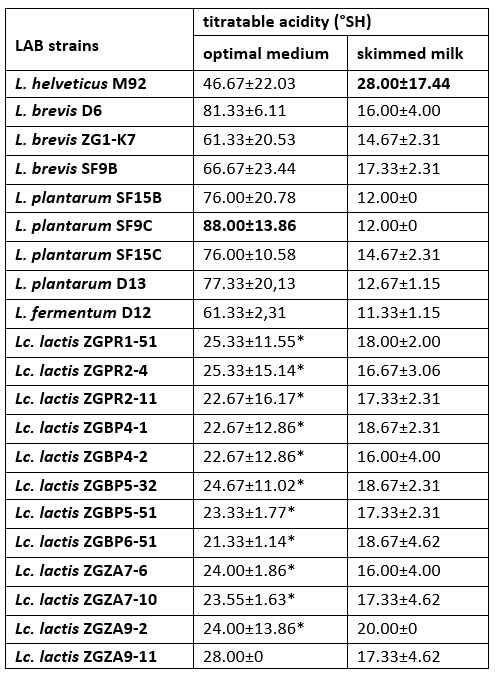
*Titratable acidities (°SH) of the culture supernatants, statistically significantly different (p<0.1) from the highest value determined in the culture supernatant of L. plantarum SF9C after overnight cultivation in the optimal medium.
Further, the potential of 21 strains of LAB to degrade casein from skimmed milk was examined (Figure 1). An easy-to-use method for determination of proteinase activity in concentrated cell biomass or in culture supernatant was used. This method preliminarily predicts the cellular localization of the proteinase by distinguishing whether it is cell wall-bound or released in the medium. For most strains, caseinolytic activity was detected in concentrated cell biomass (Figure 1). The advantage of analysing the proteolytic potential of culture supernatant or cell biomass over purified proteinase, is that it allows casein available to all components of the enzyme system. In general, the activities as observed by the appearance of transparent zones, were detected for the most of Lactobacillus strains. The obtained results indicate that the casein degradation by concentrated biomass of L. brevis D6 and L. plantarum D13 was more efficient compared to the other strains analysed (Figure 1). Strains ZGBP5-32, ZGZA7-10 and ZGZA9-2 exerted the highest proteolytic potential among lactococci. The results were confirmed both by Anson´s method and by visualization of the protein fragments resulting from casein degradation by the SDS-PAGE method (Novak et al., submitted). The differences in caseinolytic activities among the strains can be explained by the differences in molecular weight and substrate specificity of LAB proteinases.
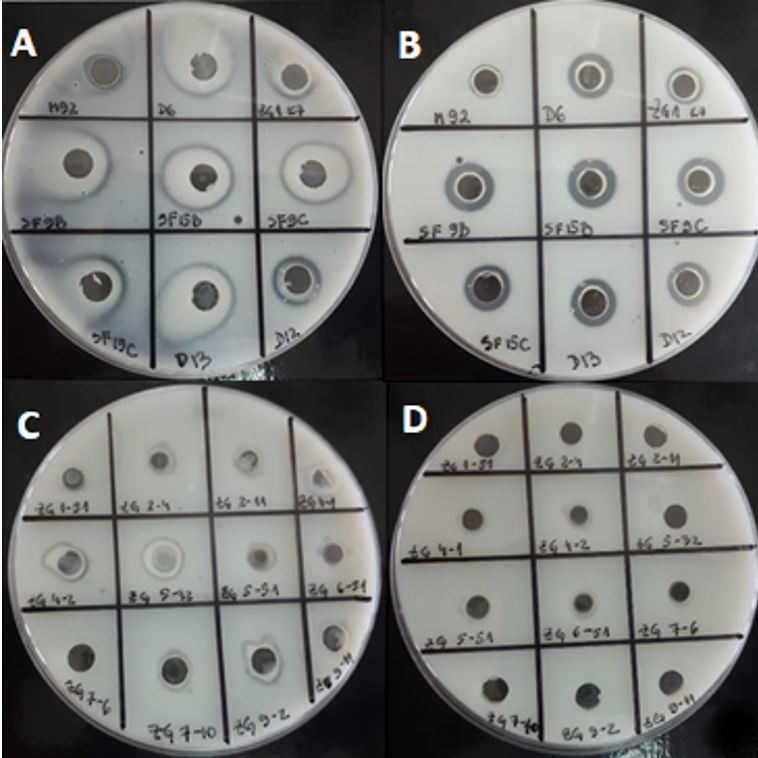
Figure 1. Transparent zones on skimmed milk agar possibly due to the proteolytic activity of LAB strains after overnight cultivation in optimal medium: A) concentrated cell biomass of 9 Lactobacillus strains B) culture supernatants of 9 Lactobacillus strains C) concentrated cell biomass of 12 Lc. lactis strains D) culture supernatants of 12 Lc. lactis strains
To assess the suitability for application in milk, LAB strains were tested in the same assay, when inoculated overnight in RSM. For certain strains the proteolytic activities were significantly changed. For example, when grown in skimmed milk, Lc. lactis ZGBP5-32, ZGBP5-51 and ZGZA9-2 and L. brevis SF9B, L. plantarum SF9C, L. helveticus M92 and L. plantarum SF15B showed the highest proteolytic activities among the tested strains (Figure 2). Interestingly, the proteolytic activity of L. plantarum SF15B, after growth in skimmed milk, was significantly higher in the sample of concentrated biomass compared to the culture supernatant. This suggests that the proteinase of L. plantarum SF15B could be bound to cell envelope. A similar was found for Lc. lactis ZGBP5-51 and ZGZA9-2 strains. Moreover, the strain Lc. lactis ZGBP5-32 showed significant proteolytic activity after overnight growth in skimmed milk.
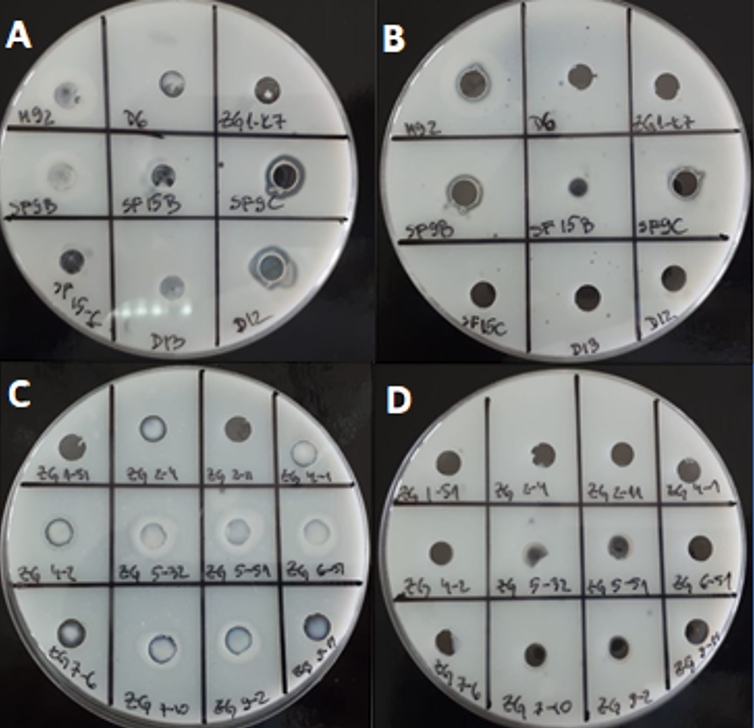
Figure 2. Transparents zones on skimmed milk agar due to the putative proteolytic activity of LAB strains after cultivation in skimmed milk: A) concentrated cell biomass of 9 Lactobacillus strains B) culture supernatants of 9 Lactobacillus strains C) concentrated cell biomass of 12 Lc. lactis strains D) culture supernatants of 12 Lc. lactis strains
To confirm casein hydrolysis, strains with the potential proteolytic activity, were surface inoculated on skimmed milk agar and incubated for 48 h at room temperature. Transparent zones of proteolysis near inoculated bacterial cultures were detected on the skimmed milk agar what confirmed caseinolytic activity of L. brevis D6, L. plantarum D13 and Lc. lactis ZGZA7-10 as illustrated in Figure 3.
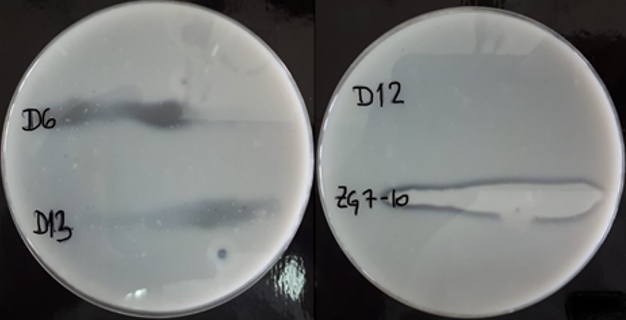
Figure 3. Transparent zones of proteolysis near the grown cultures of L. brevis D6, L. plantarum D13, L. fermentum D12 and Lc. lactis ZGZA7-10 on skimmed milk agar
Extraction of putative proteinases of Lactococcus and Lactobacillus strains
The experimental approach here is suitable for screening of proteinase expressing LAB strains, but proteinase could be futher characterised e.g. by detection of encoding enzymes through amplification of the target DNA sequences by PCR method (Leboš Pavunc et al., 2012) or by extraction of proteinase. Therefore, we used a modified non-invasive procedure of Villegas et al. (2015) who successfully performed the extraction of cell envelope proteinase, which did not disrupt the integrity of the bacterial cells and allowed extraction of proteinase with a yield of 90 %, while preserving the structure. Extraction of presumptive proteinase of 7 strains was performed, and pellets or supernatants of protein extracts were analysed by qualitative determination of proteolytic activities (Figure 4). Protein concentrations in samples of protein pellets and supernatants were determined. Their concentrations in the pellets were very low. However, the protein extracts in the pellets of the Lc. lactis strains ZGZA7-10 and ZGBP5-32 showed although low, but still detectable caseinolytic activity (Figure 4).
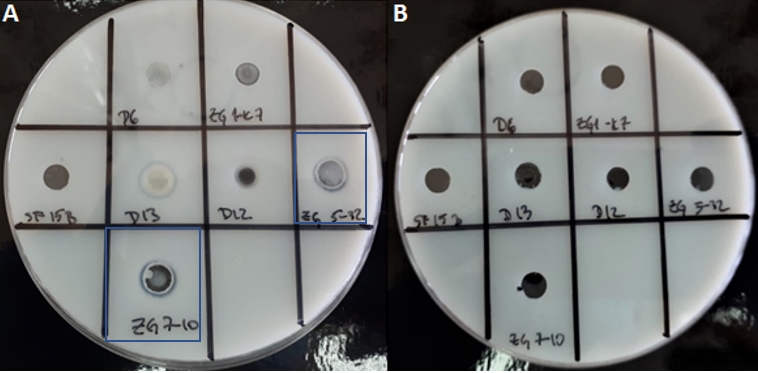
Figure 4. Zones of hydrolysis indicate the degradation of casein from skimmed milk by the crude proteinases extracts of Lc. lactis ZGZA7-10 and ZGBP5-32 in the samples of A) protein pellets, and B) supernatants of protein extracts
The putative proteinase from LAB strains was released by using CaCl 2 and obtained protein extracts were analysed by SDS-PAGE (Figure 5A and 5B). Proteins in the crude proteinase extracts of L. brevis D6, L. fermentum D12, L. plantarum D13, L. brevis SF9B and L. brevis ZG1-K7 were visualised by SDS-PAGE (Figure 5). As expected, the higher protein concentrations were measured in the samples of protein extracts which were observed as intense bands on SDS-PAGE gels (Figure 5). The protein concentrations in the concentrated cell biomass were higher compared to the concentrations in the supernatants of examined strains (data not shown).
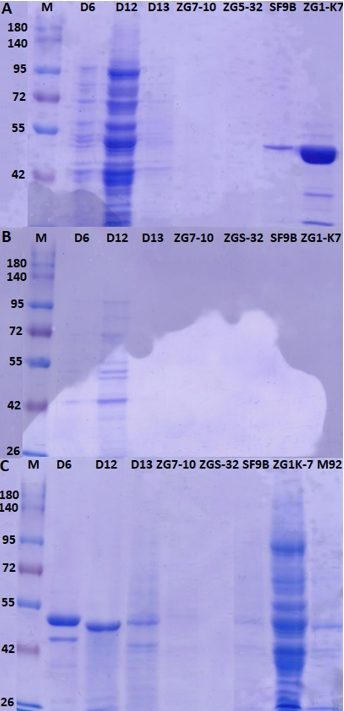
Figure 5. SDS-PAGE analysis of proteins extracts of L. brevis D6, L. fermentum D12, L. plantarum D13, Lc. lactis ZGZA7-10, Lc. lactis ZGBP5-32, L. brevis SF9B and L. brevis ZG1-K7, from samples of A) protein pellets, B) supernatants of protein extracts and C) supernatants of protein extracts after ultrafiltration
The extraction procedure is not applicable for the detection of proteins from Lc. lactis since protein bands were not detected either in the pellet sample or in the supernatant of protein extracts (Figure 5). Protein extracts of L. brevis D6, L. fermentum D12 and L. plantarum D13, contained proteins of different molecular weights isolated from either pellets or supernatants (Figure 5A and 5B), prompting a further need to optimise the protocol for removing impurities. To achieve this, released proteins were purified by an ultrafiltration membrane with a cutoff of 100 kDa. This step allowed us to remove most of the contaminating proteins, obtaining a partially purified protein extract, as observed by the presence of a single protein bands detected by SDS-PAGE (Figure 5C). Although a set of proteins was detected by SDS-PAGE, the presence of putative proteinase is still to be defined. The procedure could be further optimised by introducing different ionic strength conditions, for example by treatment with different NaCl concentrations (Villegas et al., 2015). To achieve higher yields, besides the extraction protocol, the composition of the growth medium of protease-expressing strains should be optimised. Brown et al. (2017) showed that the expression of the genes of the proteolytic system of Lactobacillus delbrueckii subsp. lactis CRL 581 was significantly lower after addition of the casein hydrolysate to the chemically defined medium. They identified a single protein band with an approximate molecular weight of 190 kDa using SDS-PAGE, which corresponds to the proteinase of the CRL 581 strain. Moreover, proteinase PrtL was not detected after producer cells were grown in MRS medium and chemically defined medium supplemented with peptides implying the importance of medium composition (Brown et al., 2017).
To summarize, the extraction should be further optimised in terms of sample purification and by a selection of cultivation media. Further research could focus on the analysis of potentially released peptides in cheese e.g. through mass spectrometry. This will establish the possible role of the enzyme in the synthesis of bioactive peptides. Efforts are also being made to develop methods for extracting bioactive peptides from the fermentation pomace (Raveschot et al., 2018). Selective precipitation with trichloroacetic acid, ethanol and ammonium sulphate is commonly used but is limited due to the additional purification steps (Agyei et al., 2016). This can be overcome by using proteolytic LAB since these bacteria produce lactic acid, which acidifies the pomace and thus may contribute to protein deposition (Raveschot et al., 2018). Finally, the use of proteolytic strains of Lactococcus or Lactobacillus, as functional starter cultures during cheese fermentation, offers an industrial applicable, low-cost approach to recover milk-derived bioactive peptides. The characterisation of the proteolytic system of probiotic bacteria contributes to the functionality of these bacteria in the context of the health effects of their biopeptides.
Conclusion
We identified proteolytically active strains of Lactobacillus and Lc. lactis isolated from Croatian fermented products. All Lc. lactis strains with the Fmc + phenotype facilitated milk coagulation, suggesting their potential application for cheese fermentation. Seven LAB, five Lactobacillus and two Lc. lactis strains were selected for further analysis based on casein proteolysis and coagulation and acidification capacities of skimmed milk. Although the SDS-PAGE analysis allowed the detection of intense bands from protein extracts of Lactobacillus strains, the procedure must be further optimised to purify the putative proteinase. Moreover, further protein identification will be performed by mass spectrometry. The characterisation of caseinolytic LAB in this study may contribute to the selection of the proteolytic strains as dairy starter cultures. Future perspectives are aimed to show whether the use of proteolytic LAB in cheese fermentation can contribute to the accumulation of peptides associated with health-promoting effects.
Funding
This work has been supported by the Croatian Science Foundation through the project IP-2019-04-2237 „Potential therapeutic biomolecules of next-generation probiotics (PRO-BIO 2.0)“ (project leader prof. Blaženka Kos). Some of the examined LAB strains were isolated within the SEE-ERA-NET Plus Project ERA-195/01 (PSALAB) and Project 0581990 Ministry of Science, Education and Sports of the Republic of Croatia (project leader prof. Jagoda Šušković).
Kazeinolitičke proteinaze Lactobacillus i Lactococcus sojeva izoliranih iz fermentiranih mliječnih proizvoda
Sažetak
Proteinaze Lactobacillus i Lactococcus sojeva kataliziraju reakcije razgradnje kazeina iz fermentiranih mliječnih proizvoda pri čemu se mogu sintetizirati peptidi s bioaktivnim učincima. Određena su proteolitička svojstva Lactococcus lactis sojeva izoliranih iz tradicionalno proizvedenih hrvatskih svježih sireva i probiotičkih te funkcionalnih starter kultura iz roda Lactobacillus. Odabrani su sojevi Lactobacillus i Lactococcus vrsta kod kojih je ustanovljen Fmc + fenotip, a koji se povezuje s prisutnošću proteinaza s kazeinolitičkim djelovanjem. Analizom fenotipa hidrolize kazeina ustanovljeno je značajnije kazeinolitičko djelovanje sojeva Levilactobacillus brevis D6 i Lactiplantilactobacillus plantarum D13 nakon kultivacije u optimalnoj hranjivoj podlozi za rast, a sojeva Lc. lactis ZGBP5-32 i Levilactobacillus brevis SF9B nakon rasta u obranom mlijeku. U svrhu procjene rasta sojeva u obranom mlijeku praćeno je zakiseljavanje mlijeka. Statistički značajna acidifikacija određena je nakon rasta L. plantarum D13 preko noći u optimalnom mediju odnosno nakon rasta proteolitičkog soja L. helveticus M92 u obranom mlijeku. Nakon pripreme ekstrakta proteina sojeva koji iskazuju kazeinolitičku aktivnost, proteinski uzorci su analizirani SDS-PAGE metodom. Djelomično pročišćene proteinaze sojeva Lc. lactis ZGBP5-32 i ZGZA7-10 i u vrlo niskim koncentracijama su pokazale kazeinolitičku aktivnost. Implementacija ultrafiltracije u postupku ekstrakcije doprinijela je pročišćavanju proteinaza što je potvrđeno SDS-PAGE metodom za proteinske ekstrakte sojeva L. brevis D6 i SF9B, L. fermentum D12 i L. plantarum D13. Daljnja istraživanja biti će usmjerena na karakterizaciju bioaktivnih peptida uslijed proteazne aktivnosti odabranih kazeinolitičkih bakterija mliječne kiseline.
Ključne riječi: bioaktivni peptidi; Fmc + fenotip; Lactobacillus; Lactococcus; proteinaze; proteolitički sustav
